Performance at any scale
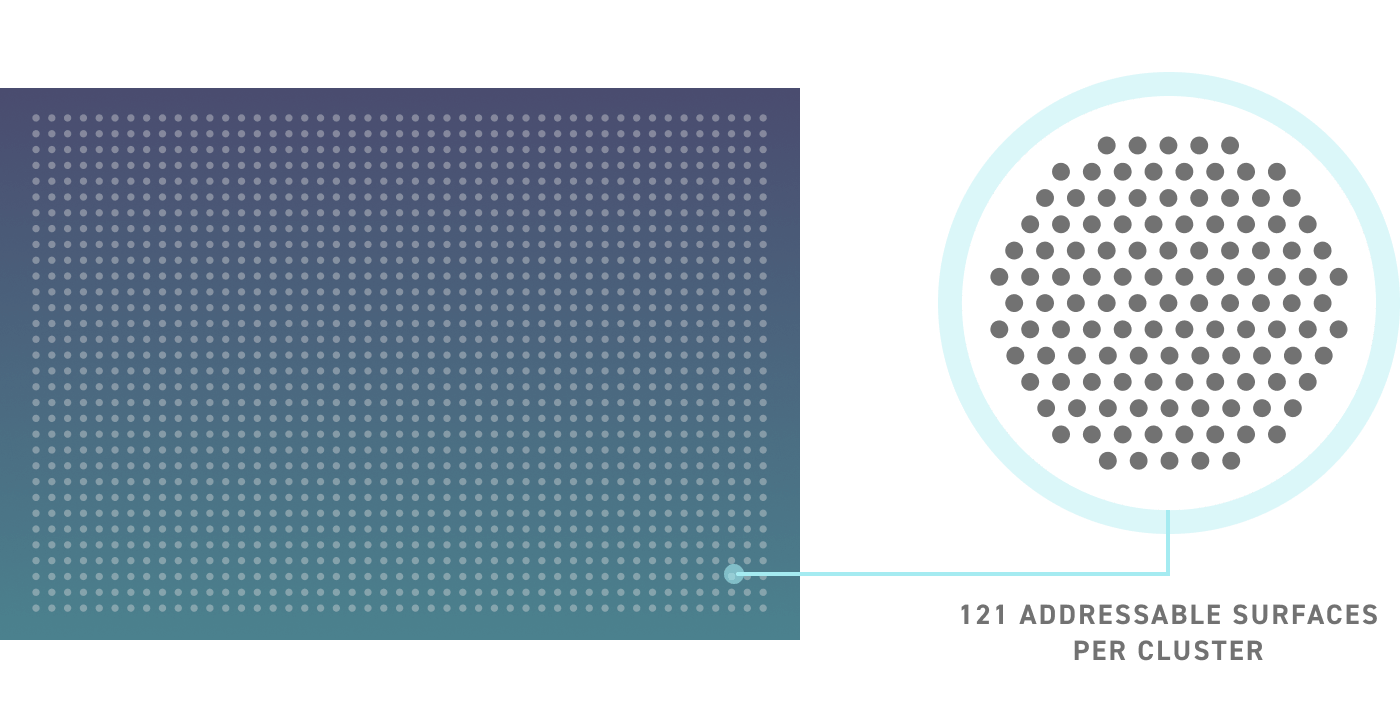
Our revolutionary silicon-based DNA Synthesis platform is capable of producing over a million unique ssDNA oligos in a single run. Each chip contains thousands of discrete clusters each containing 121 individually addressable surfaces capable of synthesizing a unique oligo sequence. A representation of the silicon chip technology can be seen below.
Due to the sheer scale of DNA synthesis capability on the chip there are virtually no limitations to the number of unique oligos that can be assembled into a pool. Don't restrict the outcome of your application, order Oligo Pools tailored precisely to your experiment.
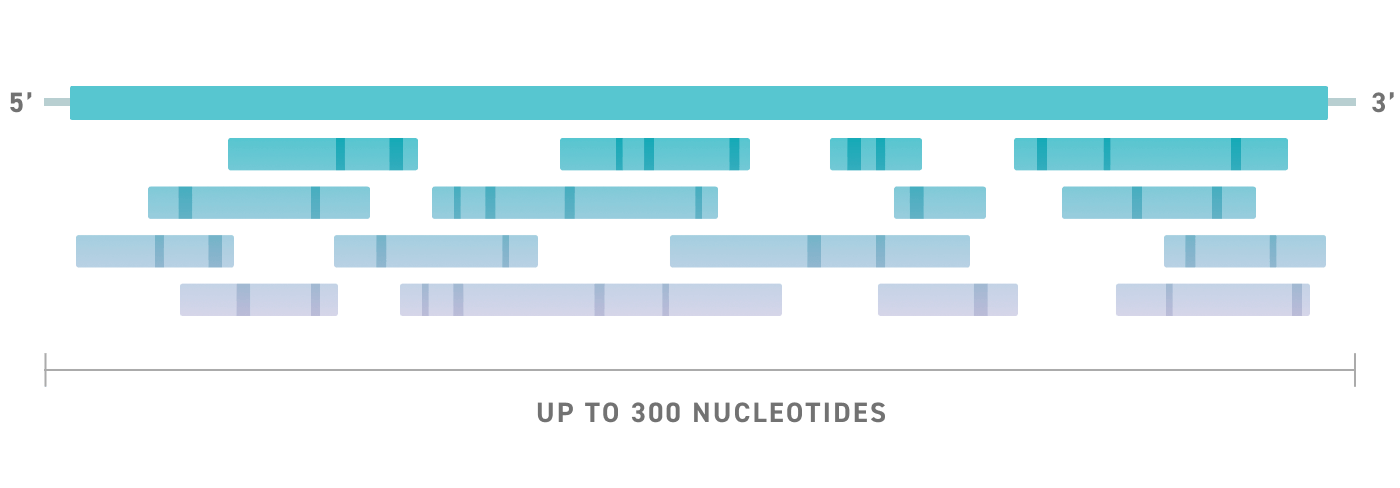
More Space for Innovation
Don’t be limited by length. Encode the elements and sequences you need, including promoters, enhancers, guides, and probes all within the expanded architecture. With oligo lengths up to 300nt it’s possible to encode multiple elements in a single design giving you the ability to do more with each oligo.
Screen once, screen right
Unmatched pool uniformity without bias provides more unique targets per screen. This means less effort is required to achieve the data coverage you need, saving time and money.
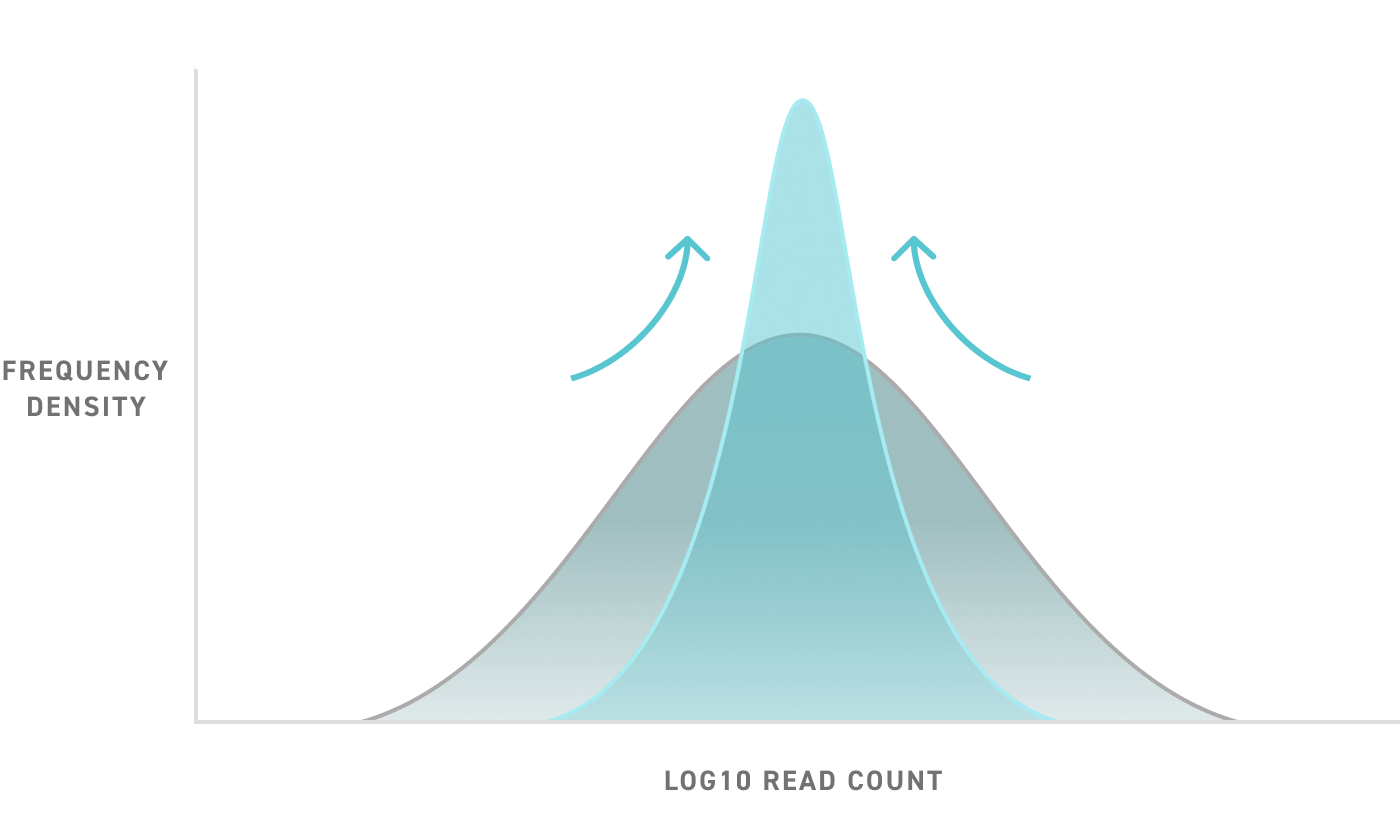
Be data confident
With the precision and uniformity of Twist Oligo Pools, you can be confident in the quality of your experimental results. Don’t miss a datapoint due to synthesis errors or inefficiencies. Get industry-leading oligo representation and low error rate for oligos of any length up to 300nt, and pools of any size. Leverage these exceptional quality oligo pools at the precise scale and length required for your application.
Tell us what you would like more information on. We’re here to help!
Contact Us
Performance at any scale
Our revolutionary silicon-based DNA Synthesis platform is capable of producing over a million unique ssDNA oligos in a single run. Each chip contains thousands of discrete clusters each containing 121 individually addressable surfaces capable of synthesizing a unique oligo sequence. A representation of the silicon chip technology can be seen below.
Due to the sheer scale of DNA synthesis capability on the chip there are virtually no limitations to the number of unique oligos that can be assembled into a pool. Don't restrict the outcome of your application, order Oligo Pools tailored precisely to your experiment.

More Space for Innovation
Don’t be limited by length. Encode the elements and sequences you need, including promoters, enhancers, guides, and probes all within the expanded architecture. With oligo lengths up to 300nt it’s possible to encode multiple elements in a single design giving you the ability to do more with each oligo.
Screen once, screen right
Unmatched pool uniformity without bias provides more unique targets per screen. This means less effort is required to achieve the data coverage you need, saving time and money.

Be data confident
With the precision and uniformity of Twist Oligo Pools, you can be confident in the quality of your experimental results. Don’t miss a datapoint due to synthesis errors or inefficiencies. Get industry-leading oligo representation and low error rate for oligos of any length up to 300nt, and pools of any size. Leverage these exceptional quality oligo pools at the precise scale and length required for your application.

Tell us what you would like more information on. We’re here to help!
Contact Us
Oligo Pools
Twist Oligo Pools are highly diverse collections of single-stranded oligonucleotides synthesized using our silicon-based DNA writing technology. Our synthesis platform enables massively parallel production of hundreds of thousands of high-quality, accurate oligos per run, allowing the generation of complex and diverse oligo pools for use in applications like CRISPR screening.
How does it work?
You provide us with your oligo sequences and we will synthesize your user designed oligo pool libraries, enabling you to spend your time working on experimentation and discovery.
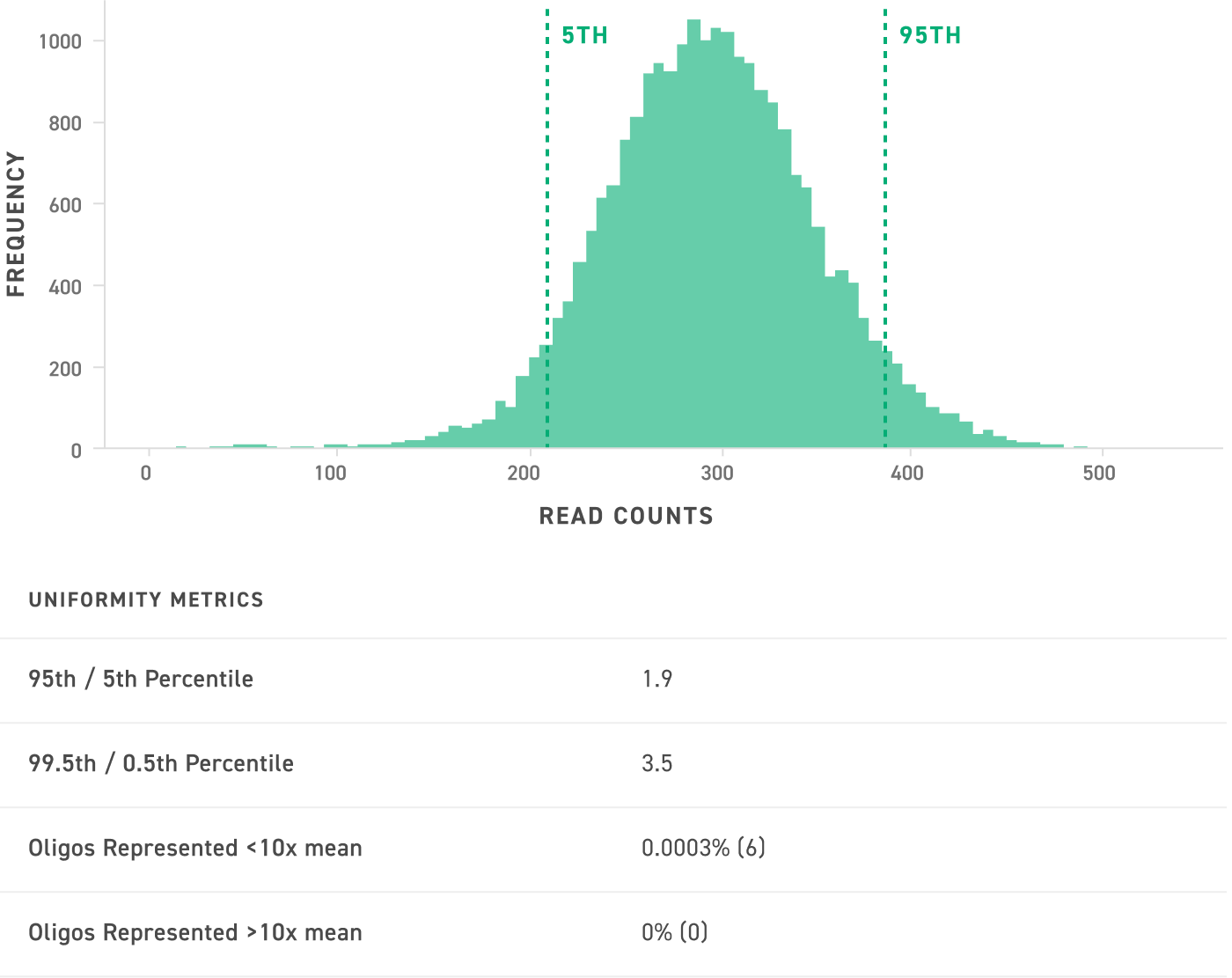
Highly Uniform Oligo Synthesis
NGS quality control data from a typical oligo pool containing 23,000 90mer oligos show the uniformity of the pool at 300x read coverage. The corresponding table indicates the uniformity metrics for this pool. Twist oligos are synthesized bias-free with high uniformity and complete oligo representation.
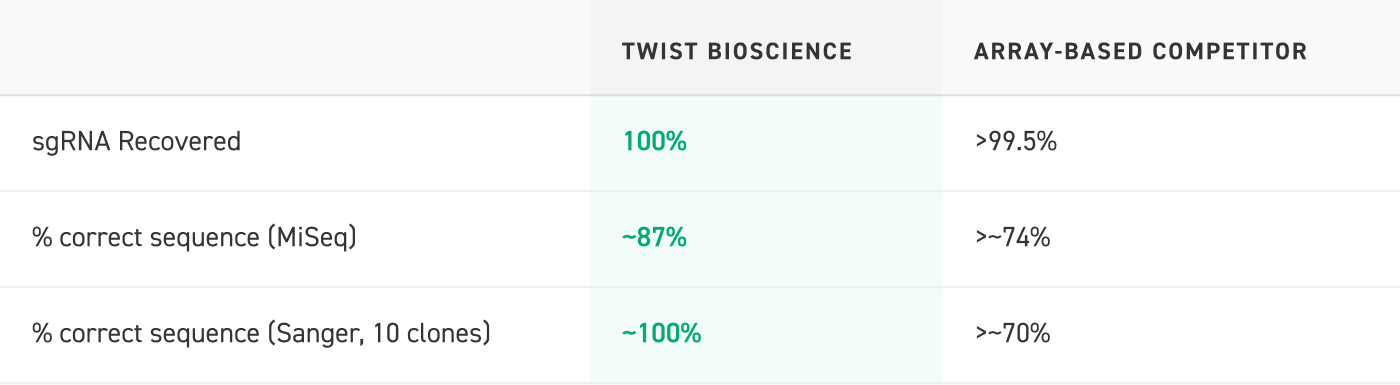
Sequencing Analysis of Oligo Pools
Sequencing analysis of Oligo Pools generated by Twist Bioscience and an array-based competitor demonstrate that the Twist Oligo Pools contain 100% of the expected sequences and a higher percentage of correct sequences than the competitor pool.
Specifications
| Oligo Length | Up to 300nt |
| Oligo Pool Size | No limits on pool size |
| Yield | >0.2 fmol average of each oligo |
| Uniformity | >90% oligos represented within <2.0x of the mean |
| Error rate | Up to 1:3000 |
| Turn-Around Time | As few as 3 days* |
| Price | Industry-leading pricing |
*Turnaround time for the standard Oligo Pool offering varies based on length and sequence complexity. Oligo Pools at 20-121 nucleotides in length range from 3-6 business days, 121-200 nucleotides in length range from 3-8 business days, and 201-300 nucleotides in length range from 5-10 business days.
Tell us what you would like more information on. We’re here to help!
Contact Us
Oligo Pools
Twist Oligo Pools are highly diverse collections of single-stranded oligonucleotides synthesized using our silicon-based DNA writing technology. Our synthesis platform enables massively parallel production of hundreds of thousands of high-quality, accurate oligos per run, allowing the generation of complex and diverse oligo pools for use in applications like CRISPR screening.
How does it work?
You provide us with your oligo sequences and we will synthesize your user designed oligo pool libraries, enabling you to spend your time working on experimentation and discovery.

Highly Uniform Oligo Synthesis
NGS quality control data from a typical oligo pool containing 23,000 90mer oligos show the uniformity of the pool at 300x read coverage. The corresponding table indicates the uniformity metrics for this pool. Twist oligos are synthesized bias-free with high uniformity and complete oligo representation.

Sequencing Analysis of Oligo Pools
Sequencing analysis of Oligo Pools generated by Twist Bioscience and an array-based competitor demonstrate that the Twist Oligo Pools contain 100% of the expected sequences and a higher percentage of correct sequences than the competitor pool.

Specifications
| Oligo Length | Up to 300nt |
| Oligo Pool Size | No limits on pool size |
| Yield | >0.2 fmol average of each oligo |
| Uniformity | >90% oligos represented within <2.0x of the mean |
| Error rate | Up to 1:3000 |
| Turn-Around Time | As few as 3 days* |
| Price | Industry-leading pricing |
*Turnaround time for the standard Oligo Pool offering varies based on length and sequence complexity. Oligo Pools at 20-121 nucleotides in length range from 3-6 business days, 121-200 nucleotides in length range from 3-8 business days, and 201-300 nucleotides in length range from 5-10 business days.
Tell us what you would like more information on. We’re here to help!
Contact Us
Cloned Oligo Pools
Oligo pool quality is the foundation of a successful experiment. Errors during synthesis or cloning can easily skew oligo pool quality, leading to over and underrepresentation of desired sequences. Twist now offers an optimized cloning service that allows you to further avoid the hardship that comes with testing PCR amplification conditions, selecting the right polymerases and primer pairs, as well as designing a cloning workflow. Skip the drama and let Twist Bioscience synthesize and clone oligo pools for you.
How does it work?
Just two steps away from your custom cloned Oligo Pools. You provide us with your oligo sequences and we will synthesize, amplify, and clone your user designed oligo pool libraries, enabling you to spend your time working on experimentation and discovery.
Twist Bioscience’s cloned oligo pools of all lengths have high uniformities and low error rates
Performance data of amplified and cloned oligo pools under 150 nucleotides in length even with high GC content:
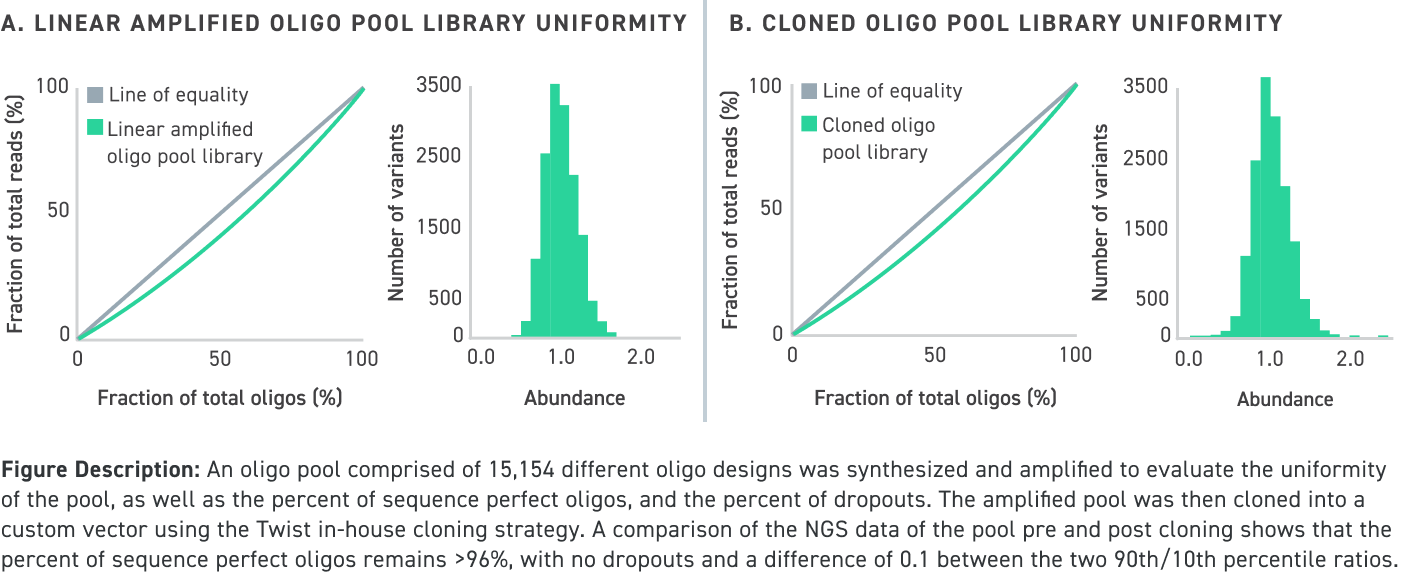
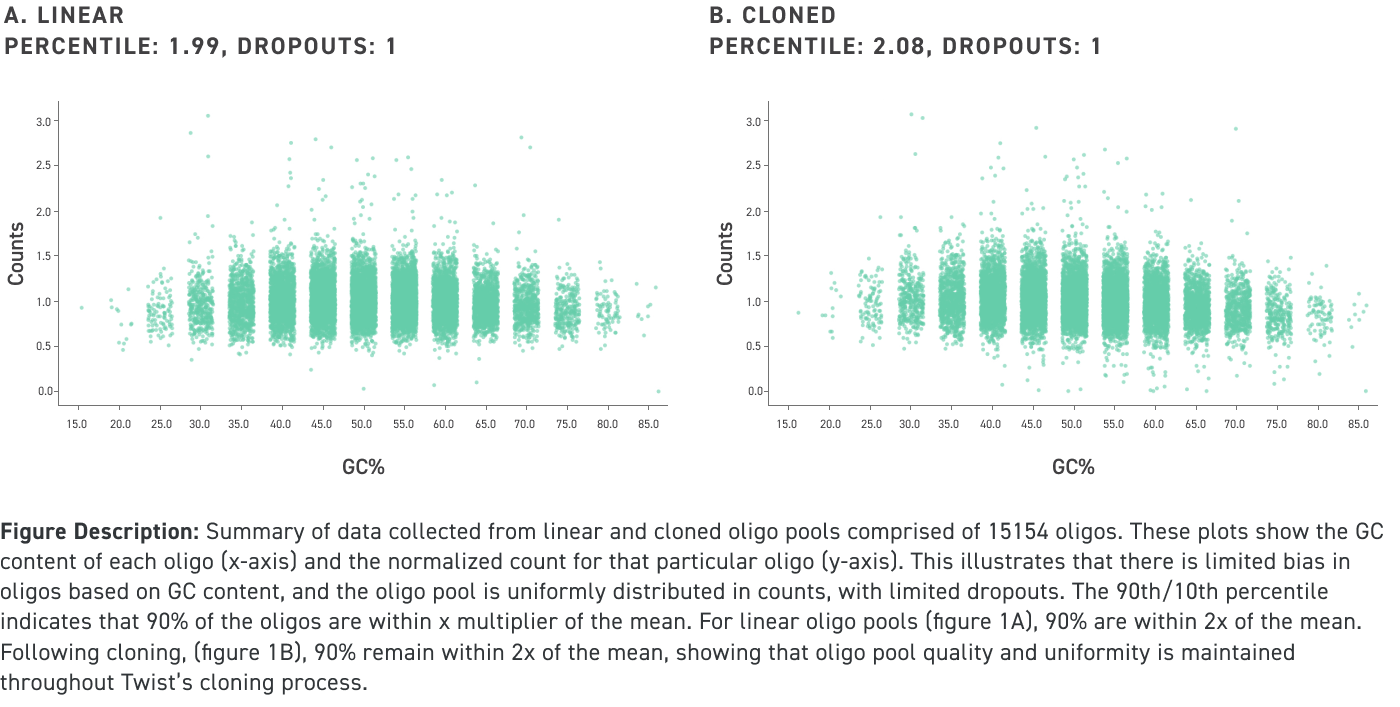
Performance data of amplified and cloned oligo pools up to 300 nucleotides in length even with high GC content:
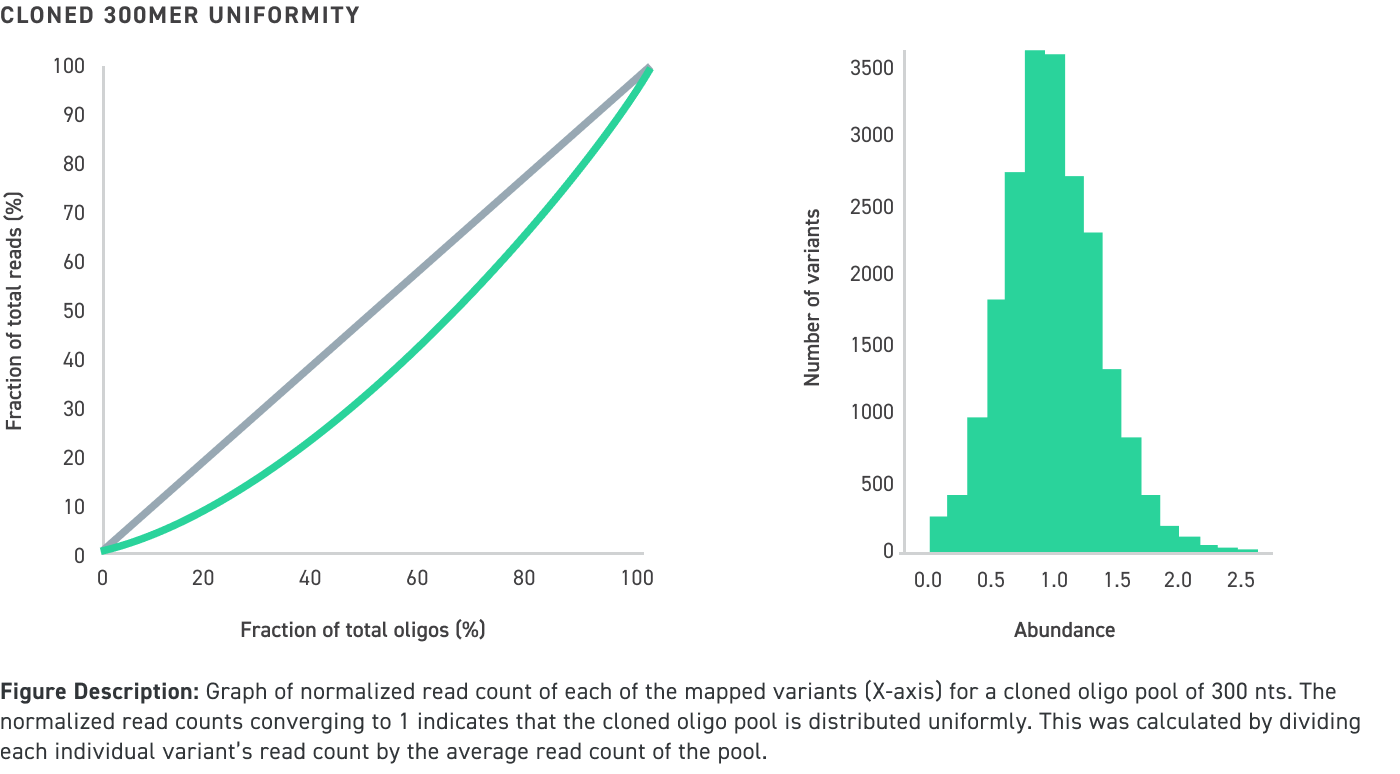
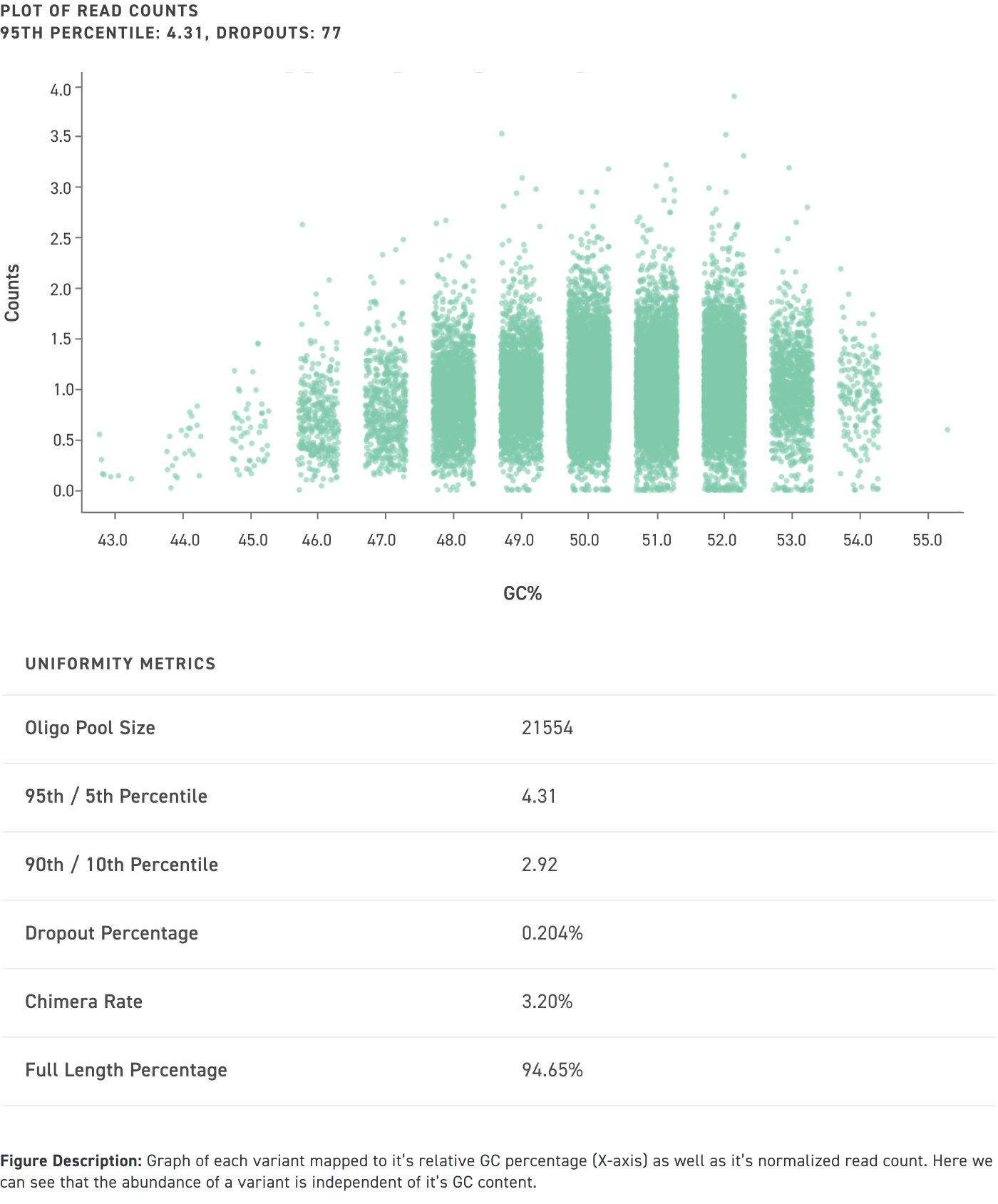
Achieve the highest quality and accuracy in your experimental data
Twist’s cloned oligo pools not only have low error rates and high uniformity, but also have low chimera rates. Chimeras are unwanted hybrid molecules that are created as a result of suboptimal PCR conditions which leads to improper amplification and recombination of different oligos within the oligo pool. See our illustration below for an example of an on-target cloned pool versus an off-target one that contains chimeras, which ultimately leads cloned oligo pools that are comprised of undesired sequences:
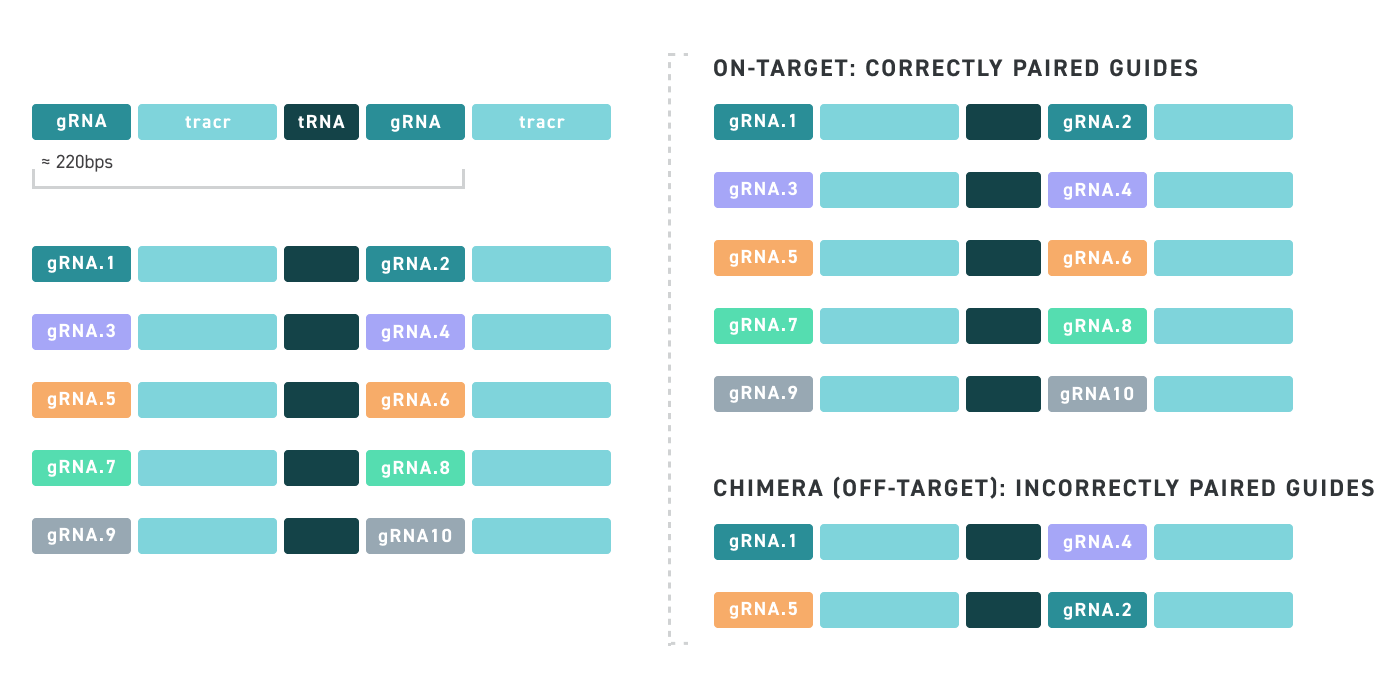
With Twist, you never have to worry about Chimeras! See for yourself:
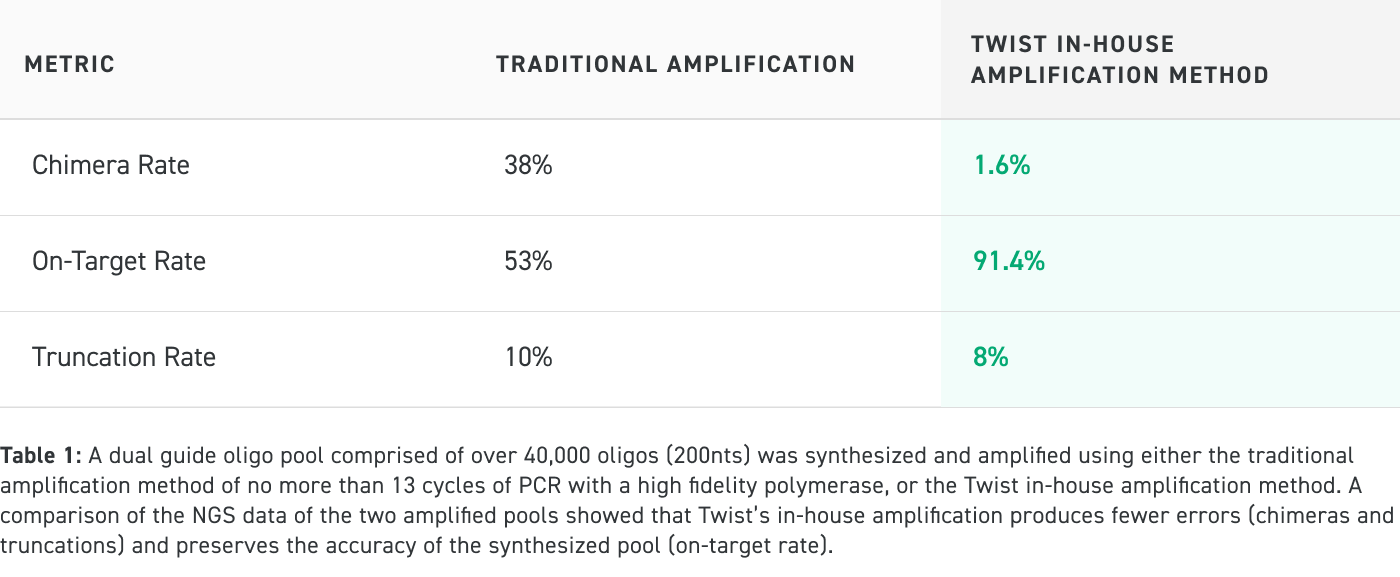
Traditional Amplification vs Twist In-House Amplification Method Performance
Specifications
| Oligo Length | Up to 300nt |
| Oligo Pool Size | No limits on pool size |
| Yield | >0.2 fmol average of each oligo |
| Uniformity | >90% oligos represented within <5.0x of the mean |
| Chimera Rate | As low as 1.5%* |
| Turn-Around Time | As few as 4 weeks* |
*Terms and Conditions: Baseline turnaround time for Cloned Oligo Pools, which includes cloning and amplification, of up to 100 and 300 nucleotides is 4-6 weeks and 6-8 weeks, respectively. An additional 2 weeks will be added to this baseline turnaround time for all Cloned Oligo Pools, regardless of length, that require new vector onboarding. Chimera rate, drop out rate, and uniformity will vary based on sequence complexity.
Tell us what you would like more information on. We’re here to help!
Contact Us
Cloned Oligo Pools
Oligo pool quality is the foundation of a successful experiment. Errors during synthesis or cloning can easily skew oligo pool quality, leading to over and underrepresentation of desired sequences. Twist now offers an optimized cloning service that allows you to further avoid the hardship that comes with testing PCR amplification conditions, selecting the right polymerases and primer pairs, as well as designing a cloning workflow. Skip the drama and let Twist Bioscience synthesize and clone oligo pools for you.
How does it work?
Just two steps away from your custom cloned Oligo Pools. You provide us with your oligo sequences and we will synthesize, amplify, and clone your user designed oligo pool libraries, enabling you to spend your time working on experimentation and discovery.

Twist Bioscience’s cloned oligo pools of all lengths have high uniformities and low error rates
Performance data of amplified and cloned oligo pools under 150 nucleotides in length even with high GC content:


Performance data of amplified and cloned oligo pools up to 300 nucleotides in length even with high GC content:


Achieve the highest quality and accuracy in your experimental data
Twist’s cloned oligo pools not only have low error rates and high uniformity, but also have low chimera rates. Chimeras are unwanted hybrid molecules that are created as a result of suboptimal PCR conditions which leads to improper amplification and recombination of different oligos within the oligo pool. See our illustration below for an example of an on-target cloned pool versus an off-target one that contains chimeras, which ultimately leads cloned oligo pools that are comprised of undesired sequences:

With Twist, you never have to worry about Chimeras! See for yourself:
Traditional Amplification vs Twist In-House Amplification Method Performance

Specifications
| Oligo Length | Up to 300nt |
| Oligo Pool Size | No limits on pool size |
| Yield | >0.2 fmol average of each oligo |
| Uniformity | >90% oligos represented within <5.0x of the mean |
| Chimera Rate | As low as 1.5%* |
| Turn-Around Time | As few as 4 weeks* |
*Terms and Conditions: Baseline turnaround time for Cloned Oligo Pools, which includes cloning and amplification, of up to 100 and 300 nucleotides is 4-6 weeks and 6-8 weeks, respectively. An additional 2 weeks will be added to this baseline turnaround time for all Cloned Oligo Pools, regardless of length, that require new vector onboarding. Chimera rate, drop out rate, and uniformity will vary based on sequence complexity.
Tell us what you would like more information on. We’re here to help!
Contact Us
Guide RNA libraries for CRISPR screens
CRISPR screens involve the precise, high-throughput knock-out, interference or modulation of gene activity, allowing complex genotype-phenotype interactions to be unveiled. Oligo Pools are designed to encode multiple guide RNA molecules per gene.
Product benefits include:
- Full control over guide library design, avoiding unwanted guides, wasted sequencing, and limited guide-per-gene ratios.
- Pairing high-fidelity Cas enzymes with highly accurate synthesis for true high-fidelity screens with minimal off-target effects.
- Incredibly uniform, bias-free pools achieve amplified signal to noise ratio to achieve maximum hit detection, lowering downstream costs.
- Oligos up to 300nt make space for additional experimental elements. Add multiple guides per oligo, genome homology, or minimal promoters with ease.
Watch our webinar: Scalable and combinatorial single-cell CRISPR screens by direct guide RNA capture and targeted seq
Genome tiling libraries
Tiling libraries allow a comprehensive study of genomic functionality. Pools of oligos may encode peptide arrays, mRNA encoding sequences, or genome fragments.
Product benefits include:
- Tiling precise genomic regions of interest. Reduce experimental noise by ignoring unwanted sequences at the design step.
- Complex, repetitive or palindromic sequences that cannot typically be synthesized as genes can be encoded in oligos.
- Larger peptide structures or genome architectures can be encoded in oligos 300nt in length.
- Uniform synthesis minimizes oversampling required for comprehensive screening.
Read our article: Identifying Coronavirus Virulence Factors for RNA Vaccine Development with Twist Oligo Pools
High throughput genetic control screens
Understanding the genetic elements controlling gene expression and regulation has broad application in synthetic biology and development of potential disease treatment. Oligo Pools power high-throughput screens of genetic control, including gene expression profiling and reporter-based applications.
Product benefits include:
- High-quality pools ensure you can interrogate every base for functionality
- High uniformity means you screen less to find your answers
- Up to 300nt pools gives you the flexibility to add multiple elements or homology regions
Watch our webinar: Epitranscriptome Screening on TechNetworks
Fluorescence in-situ hybridization
Fluorescence in-situ hybridization (FISH) allows the abundance and spatial organization of nucleic acids in biological samples to be directly visualized. Oligo Pools can encode probes that when bound to fluorescent markers provide tunable, programmable visualization of large numbers of targets.
Product benefits include:
- Rapid FISH probe development
- Sequence control enables effective targeting
- Longer oligos enable more specific targeting
Watch our webinar: Rapid and efficient production of FISH probes using oligo pools with Nicola Crosetto
Guide RNA libraries for CRISPR screens
CRISPR screens involve the precise, high-throughput knock-out, interference or modulation of gene activity, allowing complex genotype-phenotype interactions to be unveiled. Oligo Pools are designed to encode multiple guide RNA molecules per gene.
Product benefits include:
- Full control over guide library design, avoiding unwanted guides, wasted sequencing, and limited guide-per-gene ratios.
- Pairing high-fidelity Cas enzymes with highly accurate synthesis for true high-fidelity screens with minimal off-target effects.
- Incredibly uniform, bias-free pools achieve amplified signal to noise ratio to achieve maximum hit detection, lowering downstream costs.
- Oligos up to 300nt make space for additional experimental elements. Add multiple guides per oligo, genome homology, or minimal promoters with ease.
Watch our webinar: Scalable and combinatorial single-cell CRISPR screens by direct guide RNA capture and targeted seq
Genome tiling libraries
Tiling libraries allow a comprehensive study of genomic functionality. Pools of oligos may encode peptide arrays, mRNA encoding sequences, or genome fragments.
Product benefits include:
- Tiling precise genomic regions of interest. Reduce experimental noise by ignoring unwanted sequences at the design step.
- Complex, repetitive or palindromic sequences that cannot typically be synthesized as genes can be encoded in oligos.
- Larger peptide structures or genome architectures can be encoded in oligos 300nt in length.
- Uniform synthesis minimizes oversampling required for comprehensive screening.
Read our article: Identifying Coronavirus Virulence Factors for RNA Vaccine Development with Twist Oligo Pools
High throughput genetic control screens
Understanding the genetic elements controlling gene expression and regulation has broad application in synthetic biology and development of potential disease treatment. Oligo Pools power high-throughput screens of genetic control, including gene expression profiling and reporter-based applications.
Product benefits include:
- High-quality pools ensure you can interrogate every base for functionality
- High uniformity means you screen less to find your answers
- Up to 300nt pools gives you the flexibility to add multiple elements or homology regions
Watch our webinar: Epitranscriptome Screening on TechNetworks
Fluorescence in-situ hybridization
Fluorescence in-situ hybridization (FISH) allows the abundance and spatial organization of nucleic acids in biological samples to be directly visualized. Oligo Pools can encode probes that when bound to fluorescent markers provide tunable, programmable visualization of large numbers of targets.
Product benefits include:
- Rapid FISH probe development
- Sequence control enables effective targeting
- Longer oligos enable more specific targeting
Watch our webinar: Rapid and efficient production of FISH probes using oligo pools with Nicola Crosetto
Webinar
Discovering PARP inhibitor resistance mechanisms using genome-wide and focused CRISPR screens
Webinar
Discovering PARP inhibitor resistance mechanisms using genome-wide and focused CRISPR screens
Protocol
These guidelines outline the best practices for resuspending Twist Gene Fragments, Clonal Genes, Oligo Pools, and Variant Libraries.