Enhanced Methylation Detection for Liquid Biopsy Workflows
Liquid biopsy technologies are rapidly transforming the landscape of early cancer detection and monitoring. By offering a minimally invasive approach, liquid biopsies enable the analysis of circulating biomarkers such as cfDNA (cell-free DNA), which carry valuable information about cancer-associated genetic and epigenetic alterations. Among these biomarkers, DNA methylation changes stand out as a promising avenue for identifying and tracking various cancers with high specificity and sensitivity.
Despite its potential, working with cfDNA presents unique challenges due to its limited abundance and distinct fragmentation patterns. Most cfDNA fragments are approximately 167 bp in length, reflecting their nucleosome-bound origin. This fragmentation results in:
- Low stop-start diversity, resulting in more unique molecules called as duplicate sequencing reads
- Reduced library complexity, which can hinder assay sensitivity
These inherent properties make traditional methylation detection workflows less effective for cfDNA analysis, necessitating advanced tools and techniques tailored to address these limitations.
The Twist Methylated UMI Adapters enhance cfDNA methylation workflows by enabling accurate deduplication through Unique Molecular Identifiers (UMIs), reducing false duplication calls in low-diversity samples. Their design ensures compatibility with EM-Seq protocols, preserving fragment length, maximizing usable data, and resulting in improved target coverage and reproducibility—making them a critical tool for high-confidence methylation studies.
Enhanced Methylation Detection for Liquid Biopsy Workflows
Liquid biopsy technologies are rapidly transforming the landscape of early cancer detection and monitoring. By offering a minimally invasive approach, liquid biopsies enable the analysis of circulating biomarkers such as cfDNA (cell-free DNA), which carry valuable information about cancer-associated genetic and epigenetic alterations. Among these biomarkers, DNA methylation changes stand out as a promising avenue for identifying and tracking various cancers with high specificity and sensitivity.
Despite its potential, working with cfDNA presents unique challenges due to its limited abundance and distinct fragmentation patterns. Most cfDNA fragments are approximately 167 bp in length, reflecting their nucleosome-bound origin. This fragmentation results in:
- Low stop-start diversity, resulting in more unique molecules called as duplicate sequencing reads
- Reduced library complexity, which can hinder assay sensitivity
These inherent properties make traditional methylation detection workflows less effective for cfDNA analysis, necessitating advanced tools and techniques tailored to address these limitations.
The Twist Methylated UMI Adapters enhance cfDNA methylation workflows by enabling accurate deduplication through Unique Molecular Identifiers (UMIs), reducing false duplication calls in low-diversity samples. Their design ensures compatibility with EM-Seq protocols, preserving fragment length, maximizing usable data, and resulting in improved target coverage and reproducibility—making them a critical tool for high-confidence methylation studies.
Incorporating UMIs into Analysis for Improved Duplicate Resolution
UMI informed duplicate calling reduces library duplicates by over 15%, leading to an increase in library complexity (25-35%) and mean target coverage (6-8%) compared to standard positional based duplicate calling. (Figure 1)
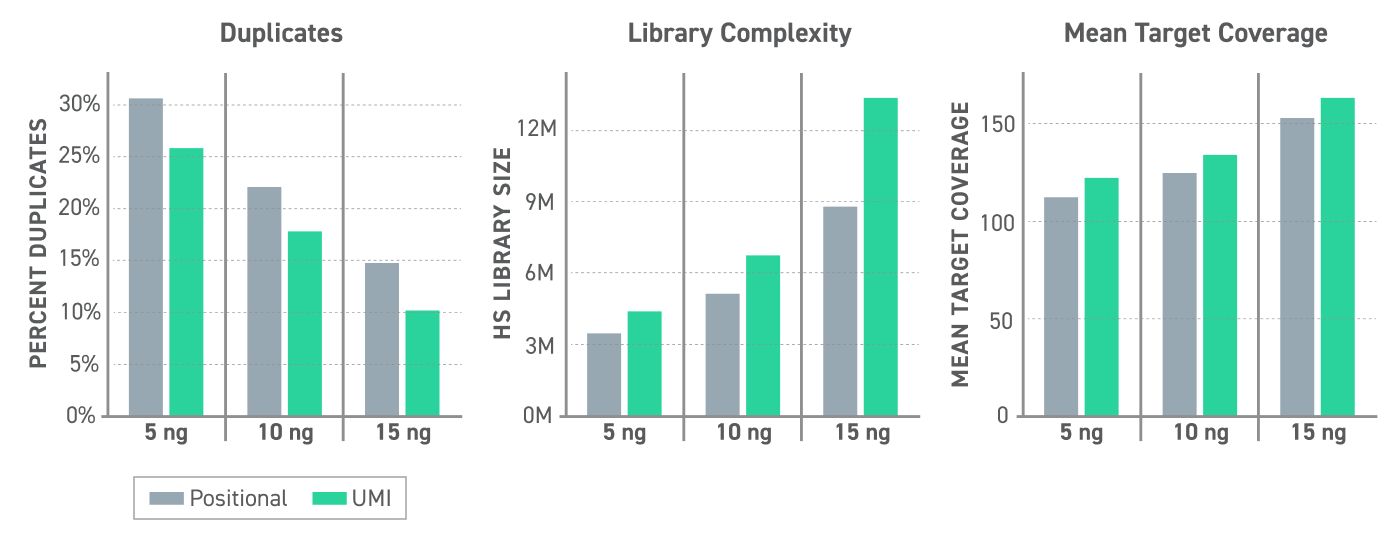
Figure 1. UMI Performance at Variable cfDNA Mass Inputs 5, 10, or 15 ng of cfDNA were used to generate EM-seq converted libraries with Twist Methylated UMI Adapters following the NEBNext EM-Seq Library Preparation. Libraries were captured with the Twist Alliance Pan-cancer Methylation Panel - 1.5MB using the Targeted Methylation Sequencing Protocol and sequenced on an Illumina Nextseq 550 to 1000X raw coverage. Duplicates were called using GATK MarkDuplicates tool (standard positional method) or called using UMIs with GATK UmiAwareMarkDuplicatesWithMateCigar tool.
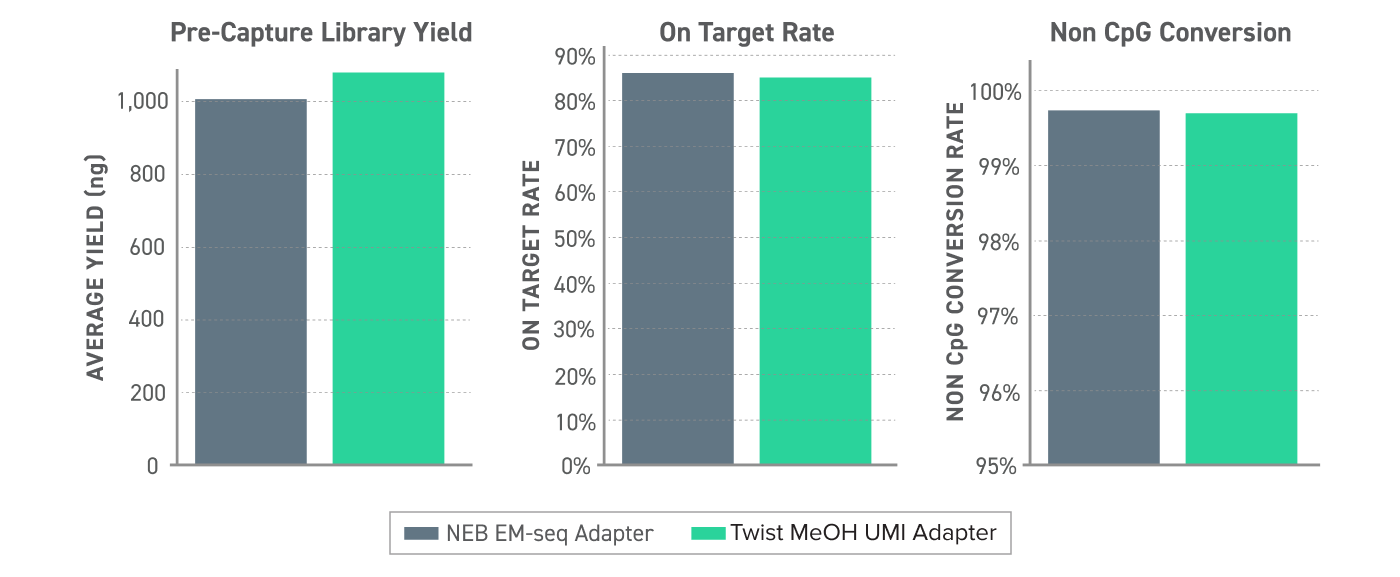
Figure 2. Methylated UMI Performance in the Twist Methylation Detection Workflow 10 ng of human cfDNA was used to generate EM-seq converted libraries with NEB EMseq adapters or the new Twist Methylated UMI Adapters. Libraries were captured with the Twist Alliance Pan-cancer Methylation Panel -1.5 MB using the Targeted Methylation Sequencing Protocol and sequenced on an Illumina Nextseq 550 to 1000X raw coverage.
Example Bioinformatics Workflow
UMI information can be integrated into a bioinformatics pipeline to remove duplicates prior to downstream analysis.

Figure 3. Summary of Analysis Workflow to Process UMIs for Dupsmarking
Raw reads are processed to mark adapter sequences and extract UMI information. Reads are then aligned to a reference genome using BWA-meth and dupsmarked using UMI information. After dupsmarking Picard metrics can be collected using GATK and additional analysis can be performed.
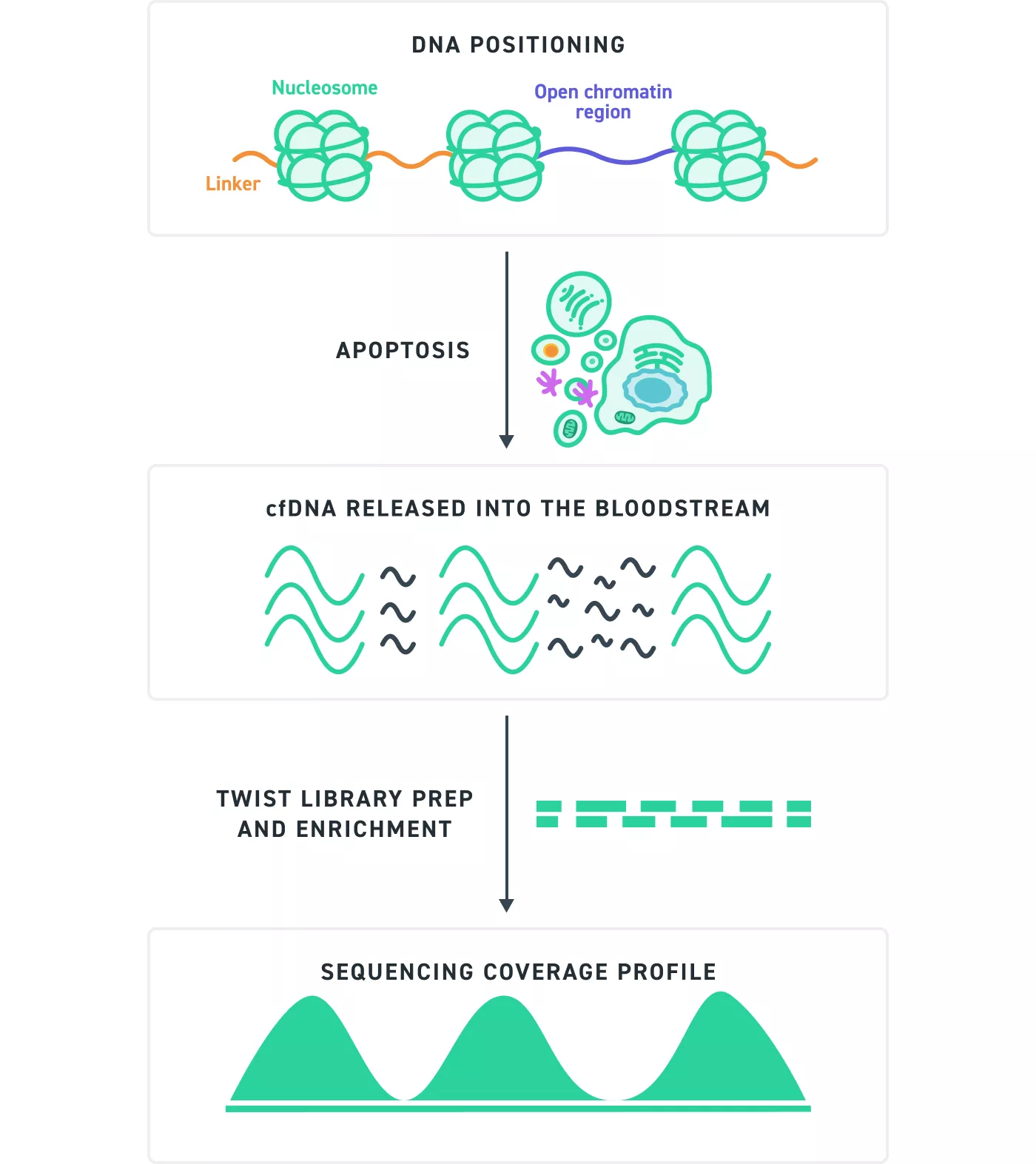
Figure 4. Schematic of Apoptotic cfDNA fragmentation to Sequencing Coverage:
During apoptosis, cell-free DNA (cfDNA) is released into the bloodstream. While endonucleases fragment DNA randomly, the way DNA is wrapped around nucleosomes introduces positional bias, leading to limited diversity in fragment start and stop sites. As a result, distinct molecules may appear as duplicates in sequencing data. Unique molecular identifiers (UMIs) are added during library preparation to label individual molecules, allowing for accurate differentiation between true duplicates and separate unique molecules.
Incorporating UMIs into Analysis for Improved Duplicate Resolution
UMI informed duplicate calling reduces library duplicates by over 15%, leading to an increase in library complexity (25-35%) and mean target coverage (6-8%) compared to standard positional based duplicate calling. (Figure 1)

Figure 1. UMI Performance at Variable cfDNA Mass Inputs 5, 10, or 15 ng of cfDNA were used to generate EM-seq converted libraries with Twist Methylated UMI Adapters following the NEBNext EM-Seq Library Preparation. Libraries were captured with the Twist Alliance Pan-cancer Methylation Panel - 1.5MB using the Targeted Methylation Sequencing Protocol and sequenced on an Illumina Nextseq 550 to 1000X raw coverage. Duplicates were called using GATK MarkDuplicates tool (standard positional method) or called using UMIs with GATK UmiAwareMarkDuplicatesWithMateCigar tool.

Figure 2. Methylated UMI Performance in the Twist Methylation Detection Workflow 10 ng of human cfDNA was used to generate EM-seq converted libraries with NEB EMseq adapters or the new Twist Methylated UMI Adapters. Libraries were captured with the Twist Alliance Pan-cancer Methylation Panel -1.5 MB using the Targeted Methylation Sequencing Protocol and sequenced on an Illumina Nextseq 550 to 1000X raw coverage.
Example Bioinformatics Workflow
UMI information can be integrated into a bioinformatics pipeline to remove duplicates prior to downstream analysis.

Figure 3. Summary of Analysis Workflow to Process UMIs for Dupsmarking
Raw reads are processed to mark adapter sequences and extract UMI information. Reads are then aligned to a reference genome using BWA-meth and dupsmarked using UMI information. After dupsmarking Picard metrics can be collected using GATK and additional analysis can be performed.
Figure 4. Schematic of Apoptotic cfDNA fragmentation to Sequencing Coverage:
During apoptosis, cell-free DNA (cfDNA) is released into the bloodstream. While endonucleases fragment DNA randomly, the way DNA is wrapped around nucleosomes introduces positional bias, leading to limited diversity in fragment start and stop sites. As a result, distinct molecules may appear as duplicates in sequencing data. Unique molecular identifiers (UMIs) are added during library preparation to label individual molecules, allowing for accurate differentiation between true duplicates and separate unique molecules.

110830
Product: Twist Methylated UMI Adapters - TruSeq Compatible, 96 Sample110830
Product: Twist Methylated UMI Adapters - TruSeq Compatible, 96 Sample