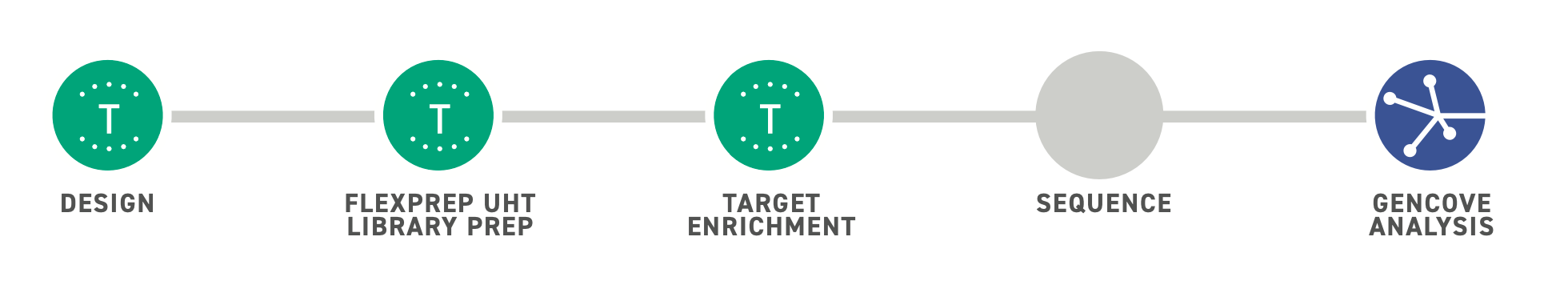
Simplified Workflow for Ultra-High Throughput Sequencing
Twist’s FlexPrepTM UHT Library Preparation Kit offers an NGS alternative to traditional microarray-based technologies for population studies, agrigenomics and other ultra-high throughput applications. Combining Twist’s Normalization by LigationTM (NBL) technology with enzymatic fragmentation methods, the kit offers built-in sample normalization across a wide range of DNA inputs, reducing both cost and complexity of typical sample processing steps. Early sample barcoding enables downstream pooling of twelve separate samples into one reaction for a more streamlined and efficient workflow.
Discover Twist's approach to ultra-high throughput sequencing



For research use only. Not for use in any diagnostic or clinical procedures.
*Based on a 8 well vs 96 well workflow
Leverage the Twist Service Lab to generate data for your next project.
See the product specifications and data behind our highest throughput library prep kit.
See how a Twist capture panel combined with FlexPrep compares to a 70k bovine array in our application note.
FAQ
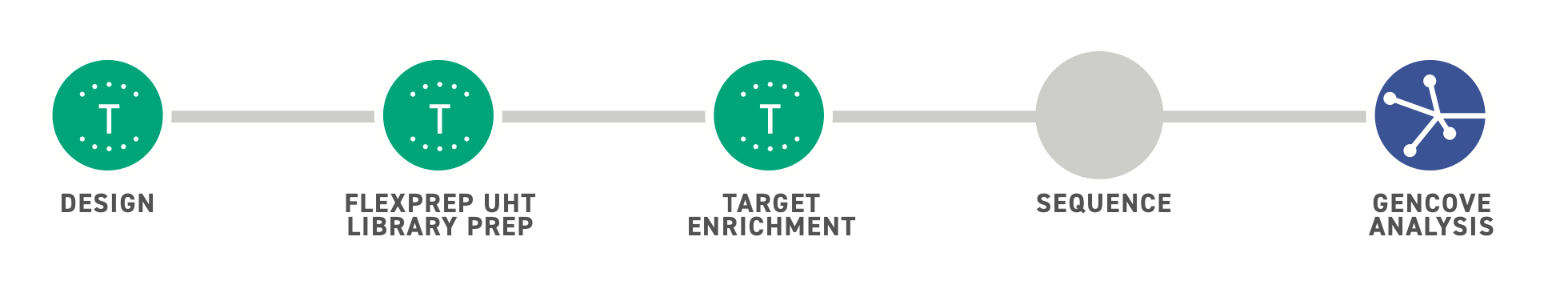
Simplified Workflow for Ultra-High Throughput Sequencing
Twist’s FlexPrepTM UHT Library Preparation Kit offers an NGS alternative to traditional microarray-based technologies for population studies, agrigenomics and other ultra-high throughput applications. Combining Twist’s Normalization by LigationTM (NBL) technology with enzymatic fragmentation methods, the kit offers built-in sample normalization across a wide range of DNA inputs, reducing both cost and complexity of typical sample processing steps. Early sample barcoding enables downstream pooling of twelve separate samples into one reaction for a more streamlined and efficient workflow.
Discover Twist's approach to ultra-high throughput sequencing



Leverage the Twist Service Lab to generate data for your next project.
See the product specifications and data behind our highest throughput library prep kit.
See how a Twist capture panel combined with FlexPrep compares to a 70k bovine array in our application note.
For research use only. Not for use in any diagnostic or clinical procedures.
*Based on a 8 well vs 96 well workflow
FAQ
The Twist FlexPrepTM UHT Library Preparation Kit takes a novel Normalization by LigationTM approach which eliminates the need for upfront and intermediate sample quantitation, streamlining your sequencing workflow.
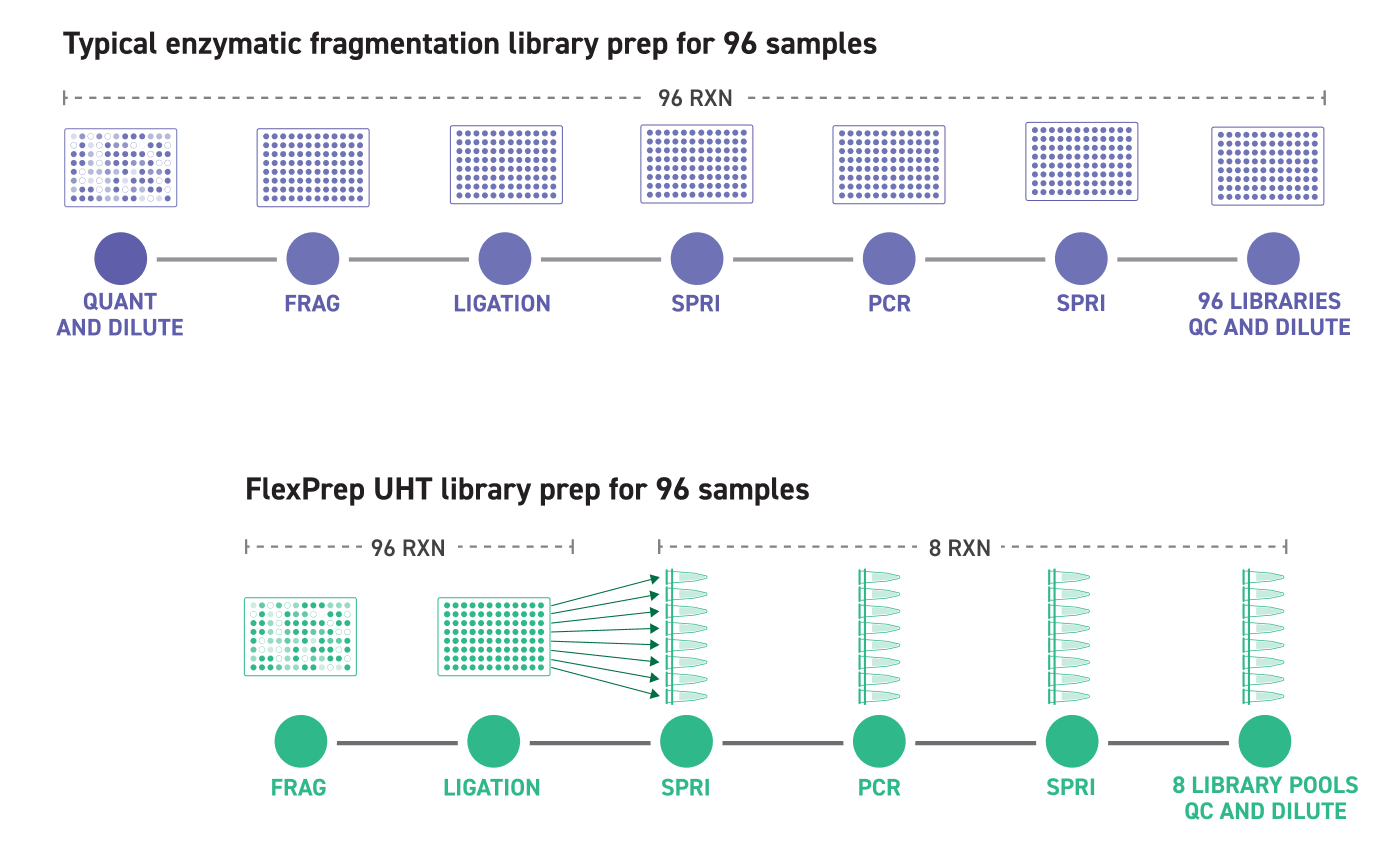
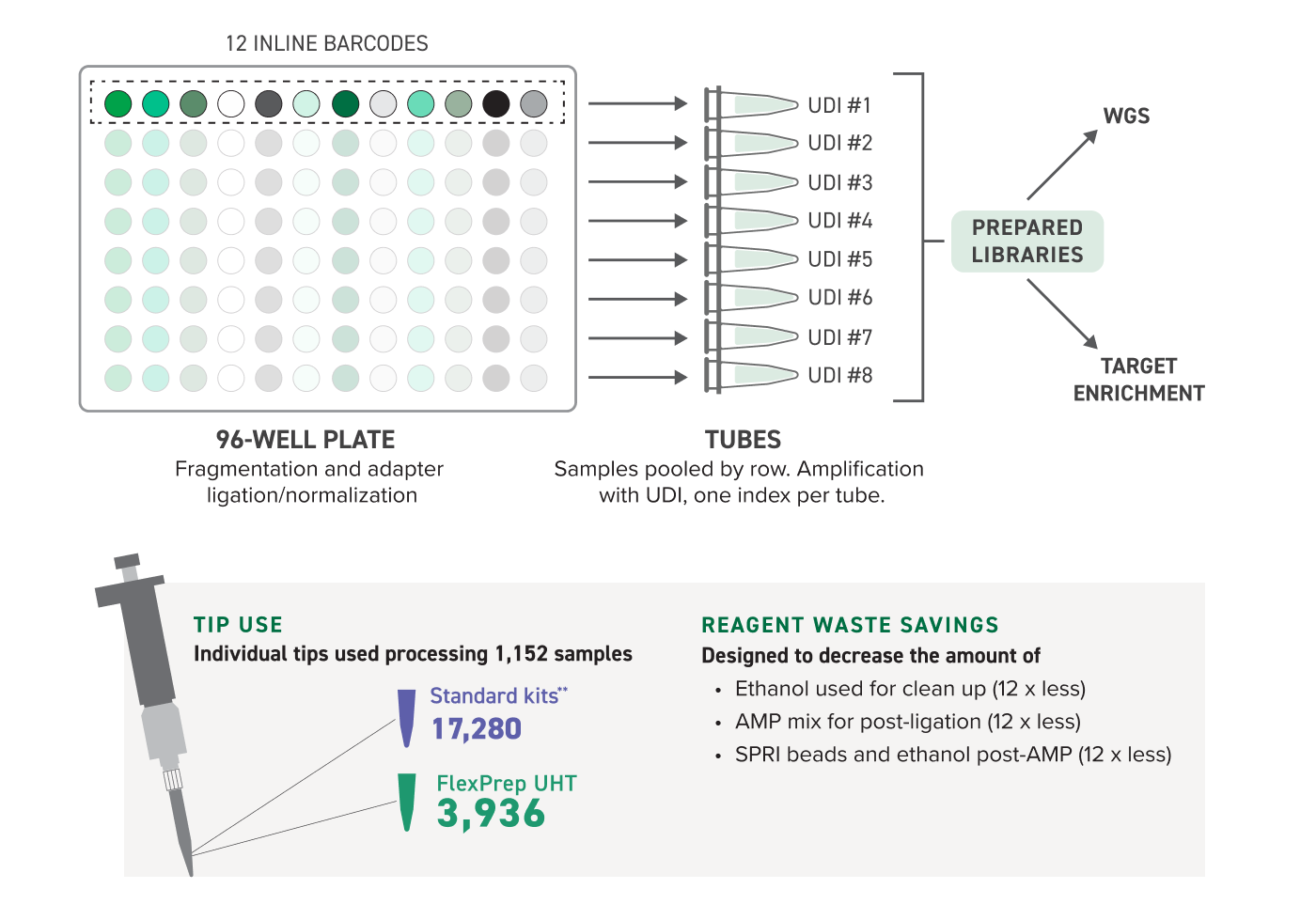
Fragmentation and ligation reactions are prepared in one well per sample. The adapters used during ligation contain inline barcodes that allow for the pooling of all 12 wells in a row of a 96-well plate. Individual pools are prepared with indices (UDIs) added by PCR for pool-level demultiplexing. Sequencing throughput can be maximized by running up to 1,152 samples in a single sequencing run from one FlexPrep kit. This increased efficiency can translate to cost and consumables savings.
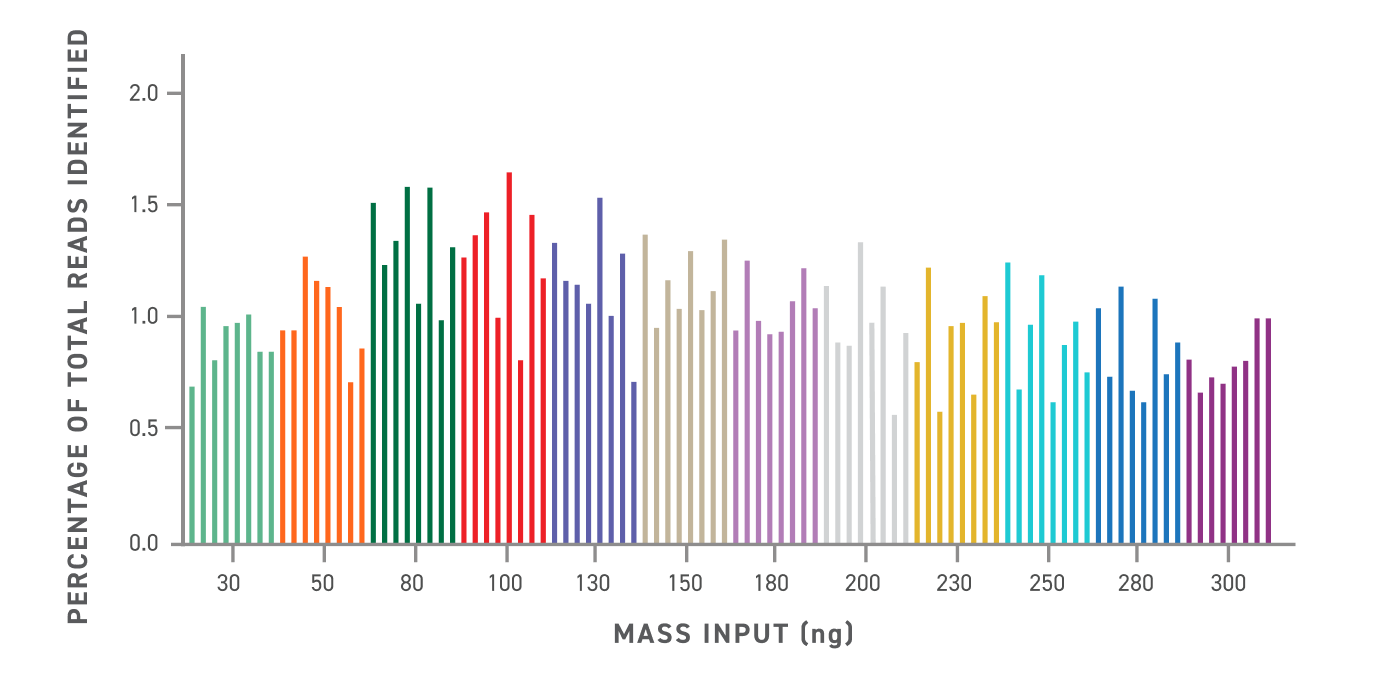
NGS read depth normalization with variable DNA mass input. Percentage of total read counts identified to each library is calculated after unique dual index and inline barcode demultiplexing. Average with perfect normalization is estimated at 1.04% (100/96).
FlexPrep UHT generates high-complexity libraries that have uniform coverage after target enrichment. In combination with Twist Custom Panels, FlexPrep offers tunable coverage of SNPs, k-mers, structural variants, and other genomic areas of interest. Enriched material was downsampled to an average of 75x coverage. Key target enrichment metrics from Picard are reported.
Target Enrichment using a 96-plex30 ng to 300 ng gDNA Mass Input into Library Preparation |
|
Metrics at 75x Raw Target Coverage |
Average +/- Standard Deviation |
Selected BasesMean Target CoverageChimerasFold-80 Base PenaltyCovered Bases at 10XCovered Bases at 20XCovered Bases at 0X |
79.22% +/- 0.54%32.74 +/- 4.781.31% +/- 0.38%1.416 +/- 0.04496.06% +/- 0.94%85.41% +/- 6.36%0.43% +/- 0.04% |
*Method used: 96 libraries were prepared with human genomic DNA (gDNA) (NA12878) using the Twist FlexPrep UHT Library Preparation Kit and FlexPrep Target Enrichment Protocol as a 96-plex with a custom 800 kb panel before sequencing on an Illumina NextSeq 550. Eight library pools of 12 samples each were generated using variable mass input ranging between 30 ng to 300 ng in each library pool. **Standard kits based on workflows which use enzymatic fragmentation followed by ligation and PCR ***All charts, figures, and graphs per Twist internal data September 2024. For research use only. Not for use in any diagnostic or clinical procedures.
The Twist FlexPrepTM UHT Library Preparation Kit takes a novel Normalization by LigationTM approach which eliminates the need for upfront and intermediate sample quantitation, streamlining your sequencing workflow.

Fragmentation and ligation reactions are prepared in one well per sample. The adapters used during ligation contain inline barcodes that allow for the pooling of all 12 wells in a row of a 96-well plate. Individual pools are prepared with indices (UDIs) added by PCR for pool-level demultiplexing. Sequencing throughput can be maximized by running up to 1,152 samples in a single sequencing run from one FlexPrep kit. This increased efficiency can translate to cost and consumables savings.

NGS read depth normalization with variable DNA mass input. Percentage of total read counts identified to each library is calculated after unique dual index and inline barcode demultiplexing. Average with perfect normalization is estimated at 1.04% (100/96).

FlexPrep UHT generates high-complexity libraries that have uniform coverage after target enrichment. In combination with Twist Custom Panels, FlexPrep offers tunable coverage of SNPs, k-mers, structural variants, and other genomic areas of interest. Enriched material was downsampled to an average of 75x coverage. Key target enrichment metrics from Picard are reported.
Target Enrichment using a 96-plex30 ng to 300 ng gDNA Mass Input into Library Preparation |
|
Metrics at 75x Raw Target Coverage |
Average +/- Standard Deviation |
Selected BasesMean Target CoverageChimerasFold-80 Base PenaltyCovered Bases at 10XCovered Bases at 20XCovered Bases at 0X |
79.22% +/- 0.54%32.74 +/- 4.781.31% +/- 0.38%1.416 +/- 0.04496.06% +/- 0.94%85.41% +/- 6.36%0.43% +/- 0.04% |
*Method used: 96 libraries were prepared with human genomic DNA (gDNA) (NA12878) using the Twist FlexPrep UHT Library Preparation Kit and FlexPrep Target Enrichment Protocol as a 96-plex with a custom 800 kb panel before sequencing on an Illumina NextSeq 550. Eight library pools of 12 samples each were generated using variable mass input ranging between 30 ng to 300 ng in each library pool. **Standard kits based on workflows which use enzymatic fragmentation followed by ligation and PCR ***All charts, figures, and graphs per Twist internal data September 2024. For research use only. Not for use in any diagnostic or clinical procedures.
FlexPrep for Human Population Genomics
With its ability to handle large cohorts efficiently and cost-effectively, FlexPrep is designed to meet the unique needs of genome-wide association studies (GWAS), genetic diversity research, and other high-throughput genomic projects.
Although many PopGen programs have readily adopted NGS technologies, new tools like FlexPrep maximize the increased throughput capabilities of today’s sequencing platforms, lowering the cost per data point.
NGS Advantages vs. Traditional Platforms for Variant Screening Studies
| Next-Generation Sequencing with FlexPrep | Standard NGS | |
|---|---|---|
| Throughput | Up to 1,152 samples in a single sequencing run; supports 96-plex enrichment | Typically processes one library per well with limited multiplexing |
| Workflow Complexity | Normalization by Ligation enables DNA input of 30-300 ng - no normalization required | More up-front and intermediary steps, including quantification, normalization |
| Consumables Usage | Sample pooling reduces tip and reagent usage 12x | Increased usage of tips and reagents lead to increased waste and costs |
Sequencing
Element Biosciences
By leveraging the Trinity Freestyle™ Hybridization Kit, FlexPrep maximizes the throughput capabilities of Element sequencers. This workflow compounds the time savings of FlexPrep’s easy workflow with the hands-off time afforded by on-sequencer Trinity enrichment chemistry.
Target Enrichment
Twist Genotyping Panel Human - 600k
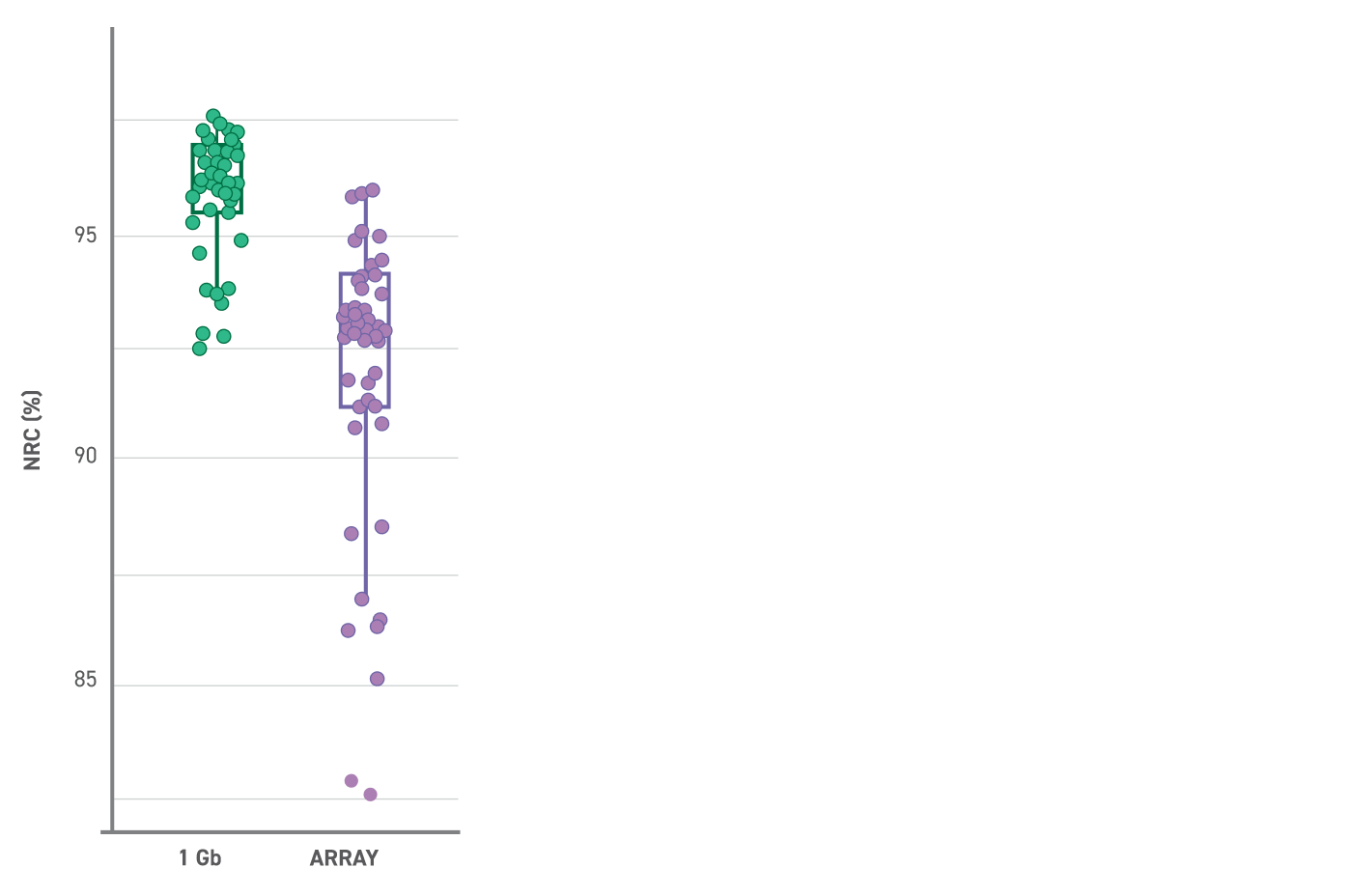
Twist Genotyping Panel - Human 600k achieves high concordance with array data. Genomewide non-reference concordance (NRC) obtained after imputing all data (sequencing or array) to the Gencove v6.1 imputation reference panel.
Analysis
Gencove
For streamlining human population genomics studies, Twist has partnered with Gencove. Combining the FlexPrep™ UHT Library Preparation Kit with Gencove's imputation pipeline enables high-confidence variant calling from reduced sequencing coverage, maximizing efficiency and throughput and allowing you to generate valuable insights from genotyping data with exceptional accuracy and reduced sequencing costs.
Leverage the Twist Service Lab to generate data for your next project.
See the product specifications and data behind our highest throughput library prep kit.
See how a Twist capture panel combined with FlexPrep compares to a 70k bovine array in our application note.
FlexPrep for Human Population Genomics
With its ability to handle large cohorts efficiently and cost-effectively, FlexPrep is designed to meet the unique needs of genome-wide association studies (GWAS), genetic diversity research, and other high-throughput genomic projects.
Although many PopGen programs have readily adopted NGS technologies, new tools like FlexPrep maximize the increased throughput capabilities of today’s sequencing platforms, lowering the cost per data point.
NGS Advantages vs. Traditional Platforms for Variant Screening Studies
| Next-Generation Sequencing with FlexPrep | Standard NGS | |
|---|---|---|
| Throughput | Up to 1,152 samples in a single sequencing run; supports 96-plex enrichment | Typically processes one library per well with limited multiplexing |
| Workflow Complexity | Normalization by Ligation enables DNA input of 30-300 ng - no normalization required | More up-front and intermediary steps, including quantification, normalization |
| Consumables Usage | Sample pooling reduces tip and reagent usage 12x | Increased usage of tips and reagents lead to increased waste and costs |
Sequencing
Element Biosciences
By leveraging the Trinity Freestyle™ Hybridization Kit, FlexPrep maximizes the throughput capabilities of Element sequencers. This workflow compounds the time savings of FlexPrep’s easy workflow with the hands-off time afforded by on-sequencer Trinity enrichment chemistry.
Target Enrichment
Twist Genotyping Panel Human - 600k
Twist Genotyping Panel - Human 600k achieves high concordance with array data. Genomewide non-reference concordance (NRC) obtained after imputing all data (sequencing or array) to the Gencove v6.1 imputation reference panel.
Analysis
Gencove
For streamlining human population genomics studies, Twist has partnered with Gencove. Combining the FlexPrep™ UHT Library Preparation Kit with Gencove's imputation pipeline enables high-confidence variant calling from reduced sequencing coverage, maximizing efficiency and throughput and allowing you to generate valuable insights from genotyping data with exceptional accuracy and reduced sequencing costs.

Leverage the Twist Service Lab to generate data for your next project.
See the product specifications and data behind our highest throughput library prep kit.
See how a Twist capture panel combined with FlexPrep compares to a 70k bovine array in our application note.
FlexPrep for Agrigenomics
In contrast to microarrays which have traditionally been used for agrigenomics studies, FlexPrep’s low cost, content flexibility, and high throughput capabilities allows for a single platform for genotyping, marker discovery, and fine mapping. When compared to standard library preparation, workflow improvements like self-normalization across sample types allows FlexPrep to tackle large agrigenomic projects like precision breeding, genomic selection, and parentage testing.
NGS Advantages vs. Traditional Platforms for Agrigenomics
| Next-Generation Sequencing with FlexPrep | Microarrays | |
|---|---|---|
| Data Completeness | Comprehensive, base-pair resolution across genome or selected regions | Limited to pre-selected variants; not well-suited for novel variant detection |
| Customization of Content | Easily customizable panels with fast TAT— target any genes, regions, or species | Content is fixed or semi-custom; changes require redesign and new manufacturing |
| Cost and Throughput | Lower cost per sample at scale due to multiplexing (up to 1152 samples per run) | Cost scales poorly for large programs or across multiple species |
Twist Workflow for Agrigenomics
Extraction
Twist DNA Purification Kit
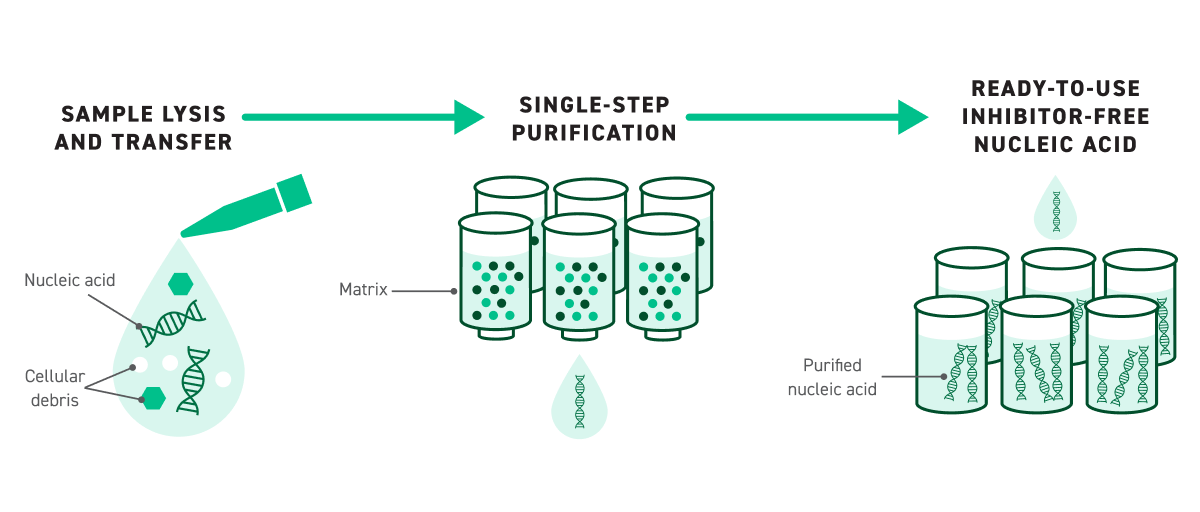
The Twist DNA Purification Kit module streamlines sample prep by taking even challenging, “dirty” samples directly into FlexPrep library preparation. Built on an innovative single-wash extraction chemistry, the kit eliminates the traditional bind–wash–elute cycle—reducing hands-on time, consumable use, and workflow complexity.
Sold as a bundled workflow with FlexPrep, the Twist FlexPrep UHT Pure Ag DNA LP Kit simplifies DNA extraction without compromising quality.
Sequencing
Element Biosciences
By leveraging the Trinity Freestyle™ Hybridization Kit, FlexPrep maximizes the throughput capabilities of Element sequencers. This workflow compounds the time savings of FlexPrep’s easy workflow with the hands-off time afforded by on-sequencer enrichment enabled by Trinity.
Analysis
Curio Genomics
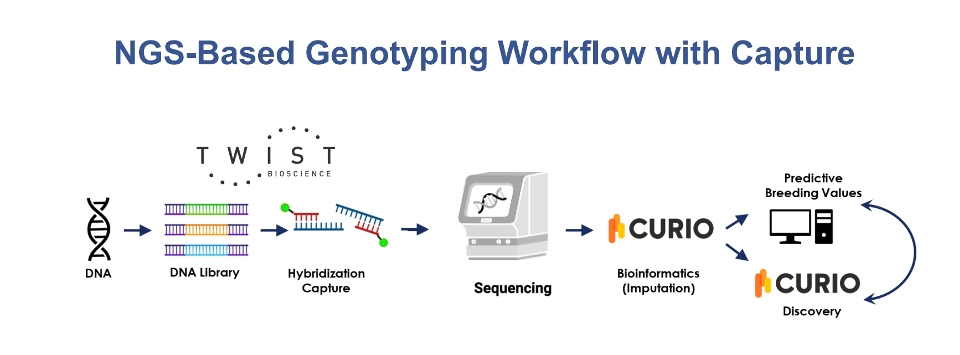
Scale your agrigenomics research with FlexPrep™ UHT Library Preparation Kit for low-pass sequencing and Curio Genomics for streamlined analysis. Twist's flexible library prep integrates seamlessly with Curio Genomics' platform, which offers high-throughput processing and efficient imputation, enabling cost-effective genotyping across large populations.
Gencove
Combining the FlexPrep™ UHT Library Preparation Kit with Gencove's imputation pipeline enables high-confidence variant calling from reduced sequencing coverage, maximizing efficiency and throughput and allowing you to generate valuable insights from genotyping data with exceptional accuracy and reduced sequencing costs.
Leverage the Twist Service Lab to generate data for your next project.
Streamline sample prep and tackle dirty samples with a seamless extraction and library prep workflow.
See how FlexPrep enables high-throughput human genotyping and compares to arrays.
See how a Twist capture panel combined with FlexPrep compares to a 70k bovine array in our application note.
FlexPrep for Agrigenomics
In contrast to microarrays which have traditionally been used for agrigenomics studies, FlexPrep’s low cost, content flexibility, and high throughput capabilities allows for a single platform for genotyping, marker discovery, and fine mapping. When compared to standard library preparation, workflow improvements like self-normalization across sample types allows FlexPrep to tackle large agrigenomic projects like precision breeding, genomic selection, and parentage testing.
NGS Advantages vs. Traditional Platforms for Agrigenomics
| Next-Generation Sequencing with FlexPrep | Microarrays | |
|---|---|---|
| Data Completeness | Comprehensive, base-pair resolution across genome or selected regions | Limited to pre-selected variants; not well-suited for novel variant detection |
| Customization of Content | Easily customizable panels with fast TAT— target any genes, regions, or species | Content is fixed or semi-custom; changes require redesign and new manufacturing |
| Cost and Throughput | Lower cost per sample at scale due to multiplexing (up to 1152 samples per run) | Cost scales poorly for large programs or across multiple species |
Twist Workflow for Agrigenomics
Extraction
Twist DNA Purification Kit
The Twist DNA Purification Kit module streamlines sample prep by taking even challenging, “dirty” samples directly into FlexPrep library preparation. Built on an innovative single-wash extraction chemistry, the kit eliminates the traditional bind–wash–elute cycle—reducing hands-on time, consumable use, and workflow complexity.
Sold as a bundled workflow with FlexPrep, the Twist FlexPrep UHT Pure Ag DNA LP Kit simplifies DNA extraction without compromising quality.

Sequencing
Element Biosciences
By leveraging the Trinity Freestyle™ Hybridization Kit, FlexPrep maximizes the throughput capabilities of Element sequencers. This workflow compounds the time savings of FlexPrep’s easy workflow with the hands-off time afforded by on-sequencer enrichment enabled by Trinity.
Analysis
Curio Genomics
Scale your agrigenomics research with FlexPrep™ UHT Library Preparation Kit for low-pass sequencing and Curio Genomics for streamlined analysis. Twist's flexible library prep integrates seamlessly with Curio Genomics' platform, which offers high-throughput processing and efficient imputation, enabling cost-effective genotyping across large populations.

Gencove
Combining the FlexPrep™ UHT Library Preparation Kit with Gencove's imputation pipeline enables high-confidence variant calling from reduced sequencing coverage, maximizing efficiency and throughput and allowing you to generate valuable insights from genotyping data with exceptional accuracy and reduced sequencing costs.

Leverage the Twist Service Lab to generate data for your next project.
Streamline sample prep and tackle dirty samples with a seamless extraction and library prep workflow.
See how FlexPrep enables high-throughput human genotyping and compares to arrays.
See how a Twist capture panel combined with FlexPrep compares to a 70k bovine array in our application note.
109220
Twist FlexPrep™ UHT Library Preparation Kit, 192 Samples109223
Twist FlexPrep™ UHT LP and Hybridization Kit, 192 Samples109224
Twist FlexPrep™ UHT Library Preparation Kit, 1152 Samples109226
Twist FlexPrep™ UHT LP and Hybridization Kit, 1152 Samples127857
Twist FlexPrep™ UHT Pure Ag DNA LP Kit, 192 Samples127858
Twist FlexPrep™ UHT Pure Ag DNA LP and Hyb Kit, 192 Samples127859
Twist FlexPrep™ UHT Pure Ag DNA LP Kit, 1152 Samples127861
Twist FlexPrep™ UHT Pure Ag DNA LP and Hyb Kit, 1152 Samples109220
Twist FlexPrep™ UHT Library Preparation Kit, 192 Samples109223
Twist FlexPrep™ UHT LP and Hybridization Kit, 192 Samples109224
Twist FlexPrep™ UHT Library Preparation Kit, 1152 Samples109226
Twist FlexPrep™ UHT LP and Hybridization Kit, 1152 Samples127857
Twist FlexPrep™ UHT Pure Ag DNA LP Kit, 192 Samples127858
Twist FlexPrep™ UHT Pure Ag DNA LP and Hyb Kit, 192 Samples127859
Twist FlexPrep™ UHT Pure Ag DNA LP Kit, 1152 Samples127861
Twist FlexPrep™ UHT Pure Ag DNA LP and Hyb Kit, 1152 Samples
Webinar
A Cost-Effective Pan-Genome-Based Platform for Genotype Imputation, QTL Mapping, and Breeding Support
Application Note
Complete and Cost-effective Agrigenomics Genotyping Across Large Breeding Populations
Application Note
Genotyping Bovine Samples Prepared With Twist FlexPrep™ Library Preparation and Target Enrichment Workflows
Application Note
Twist FlexPrep UHT Workflow Automated on Revvity Sciclone G3 NGSx Workstation
Application Note
Ultra-High Capacity Multiplexing With the Twist FlexPrep UHT Workflow Automated on Hamilton NGS STAR
Protocol
Twist FlexPrep UHT Library Preparation Kit with Enzymatic Fragmentation and Twist UDI Primers
Webinar
A Cost-Effective Pan-Genome-Based Platform for Genotype Imputation, QTL Mapping, and Breeding Support
Protocol
Twist FlexPrep UHT Library Preparation Kit with Enzymatic Fragmentation and Twist UDI Primers
Poster
Genotyping of Bovine Tissue Samples Using High-Throughput Multiplexed NGS Library Preparation Workflow
Application Note
Complete and Cost-effective Agrigenomics Genotyping Across Large Breeding Populations
Application Note
Genotyping Bovine Samples Prepared With Twist FlexPrep™ Library Preparation and Target Enrichment Workflows
Application Note
Twist FlexPrep UHT Workflow Automated on Revvity Sciclone G3 NGSx Workstation
Application Note
Ultra-High Capacity Multiplexing With the Twist FlexPrep UHT Workflow Automated on Hamilton NGS STAR