Introducing Twist Multiplexed Gene Fragments
Twist Multiplexed Gene Fragments are double stranded gene fragment sequences of up to 500 base pairs in length that are delivered as a pool instead of one fragment per well matrix. This pooled format of fragments enables high-throughput screening applications such as prime editing, ultra-complex CRISPR-based functional screening, peptide and protein engineering, Ab discovery, and massively parallel reporter assays (MPRA), where cost per fragment can be a barrier to the desired number of variants.
Multiplexed Gene Fragments are synthesized from user defined sequences in lengths of up to 500 base pairs per gene. These user-defined sequences are initially synthesized into oligo pools of up to 500 nucleotides in length and are then amplified in double-stranded DNA, and delivered as multiplexed gene fragments that have been carefully analyzed to ensure >90% of genes in the pool are the correct length. This product is optimized for rapid, high-throughput screening applications.


Testimonials
Introducing Twist Multiplexed Gene Fragments
Twist Multiplexed Gene Fragments are double stranded gene fragment sequences of up to 500 base pairs in length that are delivered as a pool instead of one fragment per well matrix. This pooled format of fragments enables high-throughput screening applications such as prime editing, ultra-complex CRISPR-based functional screening, peptide and protein engineering, Ab discovery, and massively parallel reporter assays (MPRA), where cost per fragment can be a barrier to the desired number of variants.
Multiplexed Gene Fragments are synthesized from user defined sequences in lengths of up to 500 base pairs per gene. These user-defined sequences are initially synthesized into oligo pools of up to 500 nucleotides in length and are then amplified in double-stranded DNA, and delivered as multiplexed gene fragments that have been carefully analyzed to ensure >90% of genes in the pool are the correct length. This product is optimized for rapid, high-throughput screening applications.


We ship in as few as 4 days*
Learn More

Testimonials
Twist’s Multiplexed Gene Fragment pools achieve comprehensive representation of every sequence ordered, to ensure precise control over variant construction for more targeted and rational screening.
A Multiplexed Gene Fragment pool of dual gRNA variants of 500 bp shows complete fragment representation with minimal dropouts (Fig 1) as does a pool of triple gRNA variants of 450 bp (Fig 2). Lastly, a Multiplexed Gene Fragment pool with variants containing GC content across a wide range including fragments with up to 80% GC shows high representation and minimal bias (Fig 3).
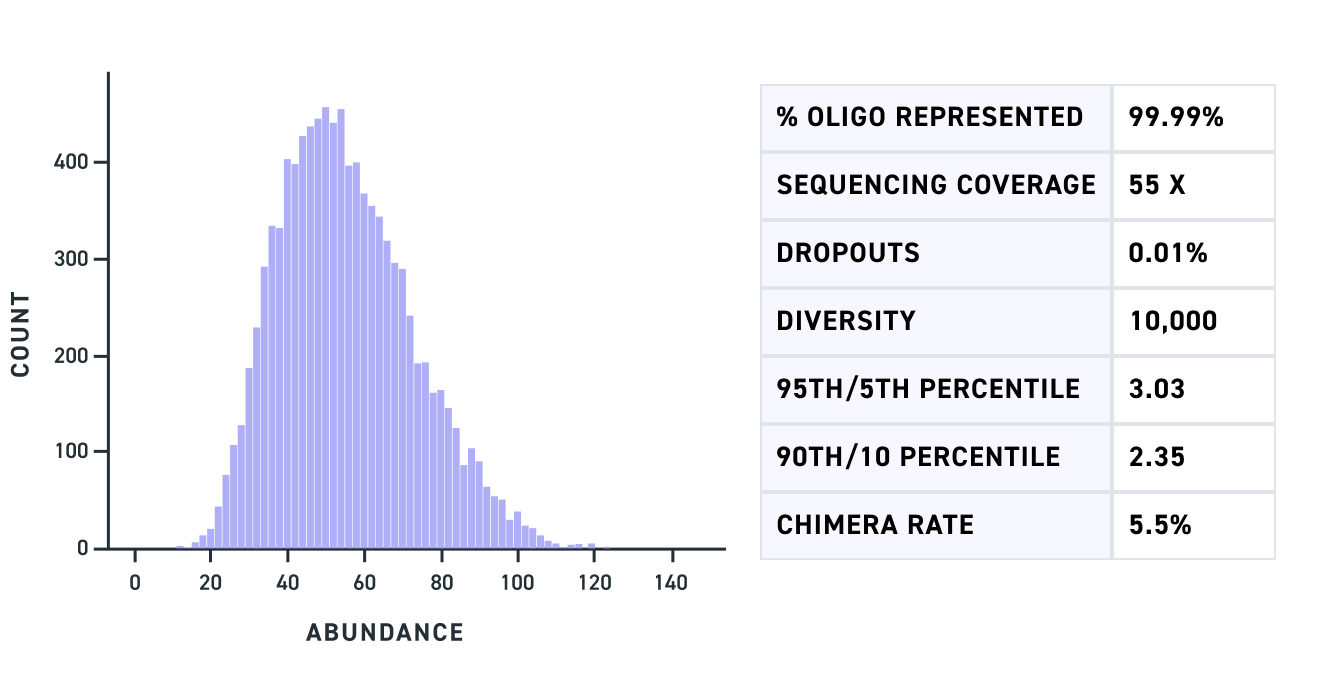
Figure 1. Graph of read count of each of the mapped dual gRNA variants (X-axis) for an amplified pool of Multiplexed Gene Fragments of 500 bp.
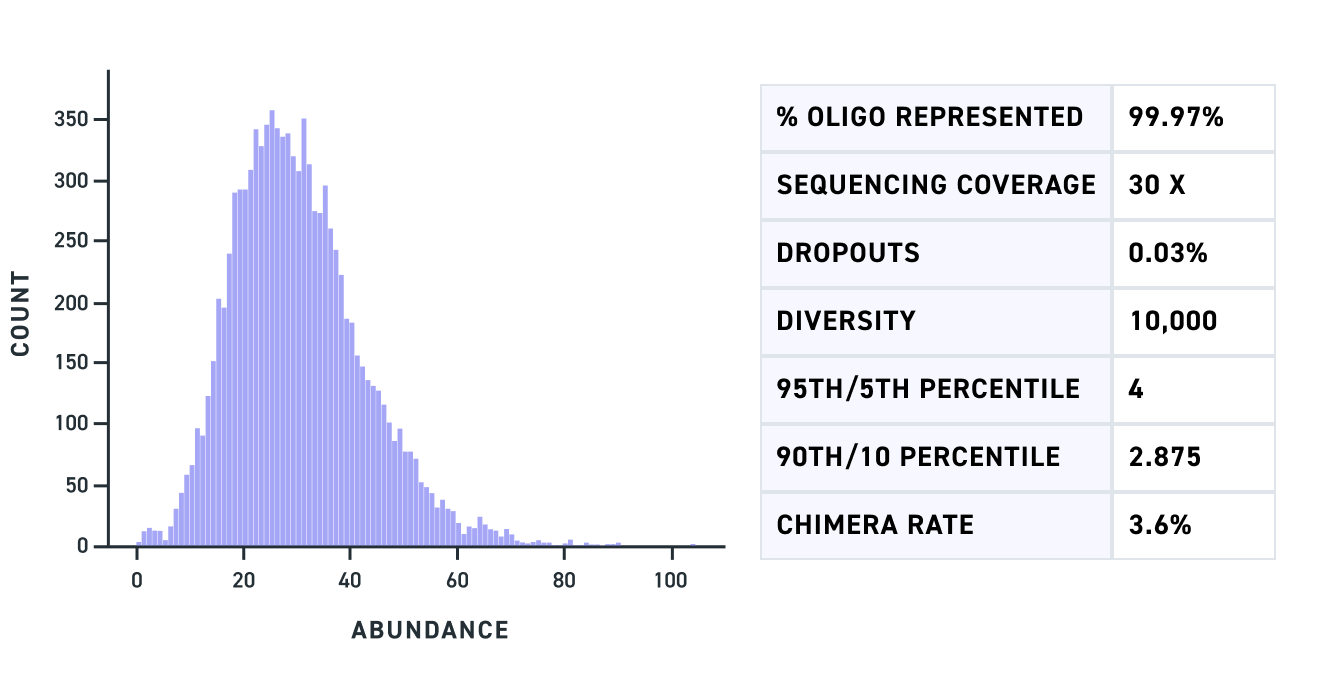
Figure 2. Graph of read count of each of the mapped triple gRNA variants (X-axis) for an amplified pool of Multiplexed Gene Fragments of 450 bp.
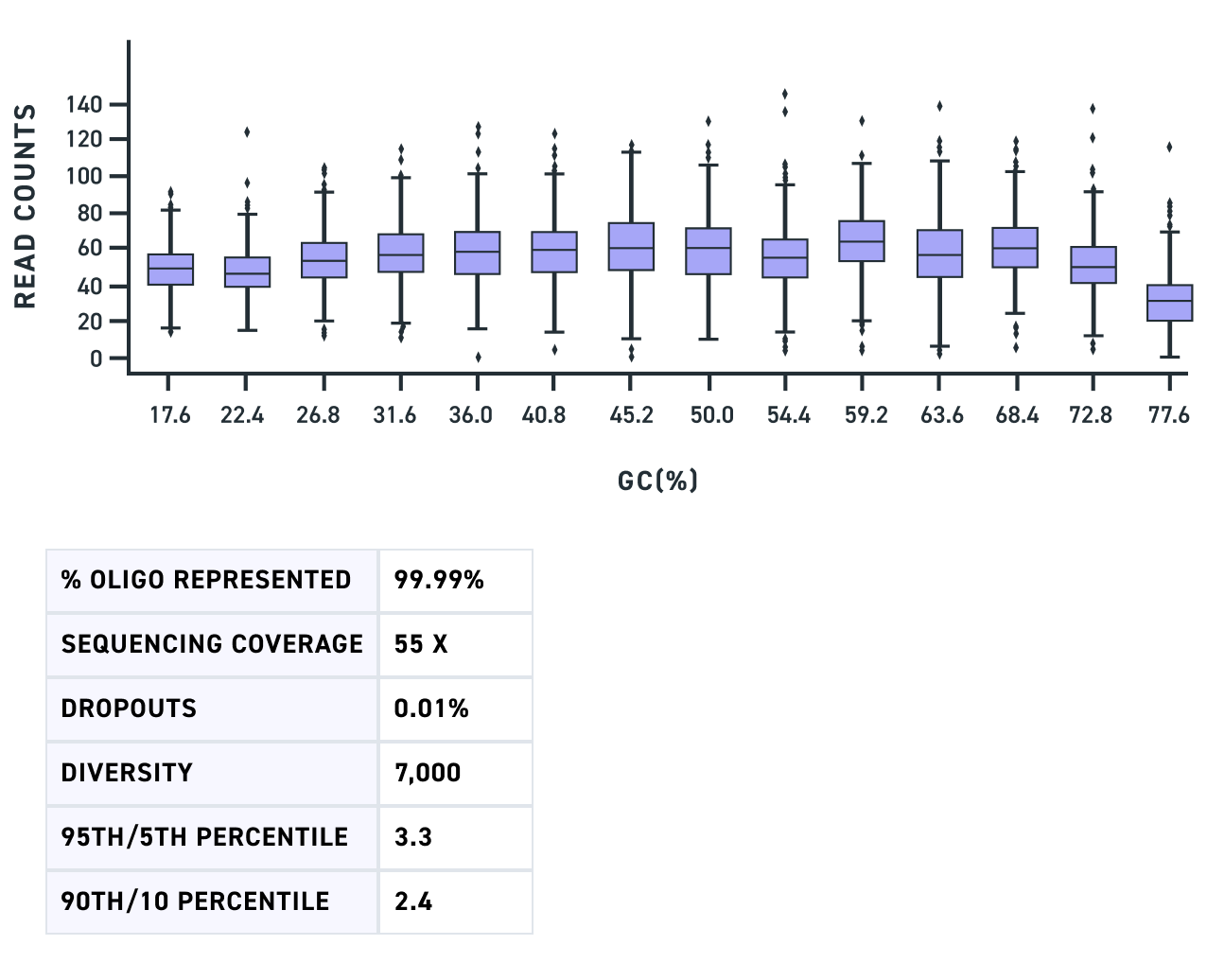
Figure 3. GC plots of an amplified pool of Multiplexed Gene Fragments of 500 bp length were created and show minimal bias among a wide range of GC content up until 80%. X-axis are categories of 14 groups of sequences with different GC content. Y-axis is the read count.
Twist’s Multiplexed Gene Fragment pools achieve comprehensive representation of every sequence ordered, to ensure precise control over variant construction for more targeted and rational screening.
A Multiplexed Gene Fragment pool of dual gRNA variants of 500 bp shows complete fragment representation with minimal dropouts (Fig 1) as does a pool of triple gRNA variants of 450 bp (Fig 2). Lastly, a Multiplexed Gene Fragment pool with variants containing GC content across a wide range including fragments with up to 80% GC shows high representation and minimal bias (Fig 3).

Figure 1. Graph of read count of each of the mapped dual gRNA variants (X-axis) for an amplified pool of Multiplexed Gene Fragments of 500 bp.

Figure 2. Graph of read count of each of the mapped triple gRNA variants (X-axis) for an amplified pool of Multiplexed Gene Fragments of 450 bp.

Figure 3. GC plots of an amplified pool of Multiplexed Gene Fragments of 500 bp length were created and show minimal bias among a wide range of GC content up until 80%. X-axis are categories of 14 groups of sequences with different GC content. Y-axis is the read count.
Yield
Minimum of 200 ng of amplified dsDNATurnaround time
8-12 business daysDelivery Format
Dried-down, dsDNA pooled in a 2 mL tubeLength
301 - 500 bpPool size
No minimum, no maximumUniformity
90% of sequences are within ~3x of the mean*Quality Control (QC)
Fragment analysis to ensure >90% of genes in a pool are the correct length*Error Rate
1:2000 nt (from the original pool synthesis, before amplification)*Terms and Conditions: Standard turnaround time for Multiplexed Gene Fragments is 8-12 business days. This will vary based on sequence length and complexity. Multiplexed Gene Fragment length is up to 500 base pairs, with up to 460 bp allotted for the variable region with a minimum of 40 bp conserved for the primer region. Quality control tests ensure >90% of gene sequences within a pool will be user defined length.
For research use only. Not for Diagnostic Procedures.
Yield
Minimum of 200 ng of amplified dsDNATurnaround time
8-12 business daysDelivery Format
Dried-down, dsDNA pooled in a 2 mL tubeLength
301 - 500 bpPool size
No minimum, no maximumUniformity
90% of sequences are within ~3x of the mean*Quality Control (QC)
Fragment analysis to ensure >90% of genes in a pool are the correct length*Error Rate
1:2000 nt (from the original pool synthesis, before amplification)*Terms and Conditions: Standard turnaround time for Multiplexed Gene Fragments is 8-12 business days. This will vary based on sequence length and complexity. Multiplexed Gene Fragment length is up to 500 base pairs, with up to 460 bp allotted for the variable region with a minimum of 40 bp conserved for the primer region. Quality control tests ensure >90% of gene sequences within a pool will be user defined length.
For research use only. Not for Diagnostic Procedures.
Let's Get Started
- Step 1: Complete the contact details form on this page
- Step 2: Download the MGF Submission Form and fill out all fields in the form for your MGF design
- Step 3: Upload the completed submission form to the "Submit File" tab on this page
If you have any questions, please feel free to email us at customersupport@twistbioscience.com
Ready to Submit?
- Step 1: Upload completed submission form
- Step 2: Our MGF Team Experts will review your project
- Step 3: MGF Team verifies the project and will reach out to you with a quote
- Step 4: Once the quote is accepted by the customer, MGF Team will send the project to the production team
If you have any questions, please feel free to email us at customersupport@twistbioscience.com
Let's Get Started
- Step 1: Complete the contact details form on this page
- Step 2: Download the MGF Submission Form and fill out all fields in the form for your MGF design
- Step 3: Upload the completed submission form to the "Submit File" tab on this page
If you have any questions, please feel free to email us at customersupport@twistbioscience.com
Ready to Submit?
- Step 1: Upload completed submission form
- Step 2: Our MGF Team Experts will review your project
- Step 3: MGF Team verifies the project and will reach out to you with a quote
- Step 4: Once the quote is accepted by the customer, MGF Team will send the project to the production team
If you have any questions, please feel free to email us at customersupport@twistbioscience.com