Exome Sequencing 101: Part 1 - Library Preparation
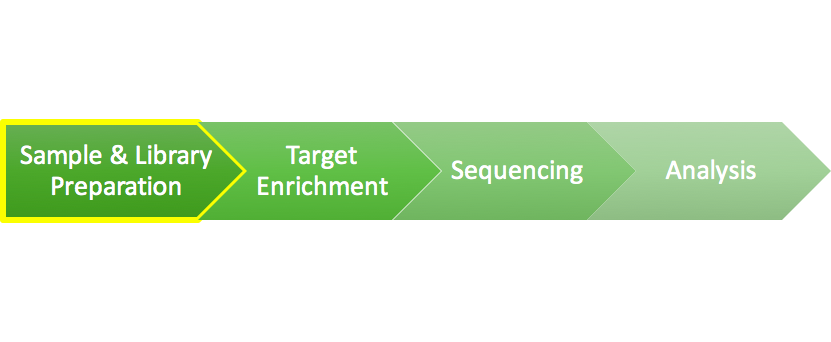
Rapidly decreasing DNA sequencing costs are driving improvements in personalized medicine. Refining and analyzing just the regions that encode our genes when sequencing a person’s genome will both lower costs and increase accessibility of these treatments. But what does the exome sequencing process look like in the lab? This piece is the first of four parts to demystify the exome sequencing process.
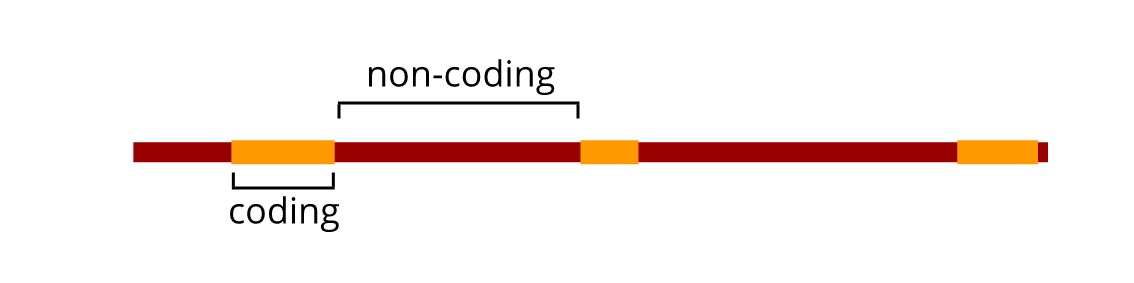
A human genome is incredibly complex. If you look at the genes (the parts of the genome that encode proteins), you will see that they are fragmented, scattered across the genome, surrounded by other DNA. That other DNA is required for maintaining the genome’s integrity, for controlling each gene’s expression, and in some instances, its function still remains a mystery. A researcher’s aim in an exome sequencing experiment is to isolate the DNA sequences from a genomic sample containing only the protein coding regions. This is the exome. Only 1% of a human genome contains gene encoding regions[1], yet around 85% of genetic mutations known to cause disease occur in the exome[2]. By isolating just these regions, the amount of genomic DNA that needs to be sequenced to get meaningful data about a disease can be lowered. This provides an important, low cost “first pass” screen for mutations.
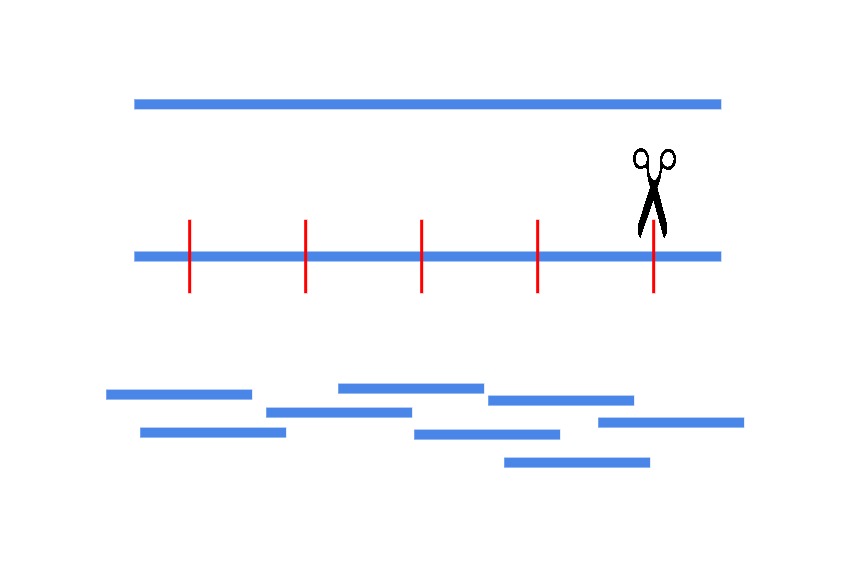
Next generation sequencing platforms often make DNA reads only a few hundred letters long, however they can scan millions of short pieces of DNA at once. Then the researcher can add the pieces back into the full genome picture using specialized software. A researcher therefore needs to break a patient’s genomic sample into millions of short length fragments. This is the fragment library, and its preparation is the first step in any exome sequencing experiment.
Let’s use a clinical scenario as an example:
A patient has been diagnosed with early onset alzheimer's disease, and as is often the case with rare inherited diseases, the patient’s doctor would like to get a deeper understanding of the disease’s underlying cause in the patient’s genome. Doing so may both advance science’s understanding of this particular disease, and it may help the doctor plan a targeted therapy that will give the patient the best possible prognosis.
In this scenario, the doctor might contact a company who specializes in genetic testing and order an exome sequencing experiment to focus in on the regions that are already known or suspected to contain the disease-causing mutations.
A cell sample is then taken from the patient, and sent to a research laboratory for sequencing. A researcher takes the cells and performs an extraction of the genomic DNA—simple kits are available on the market to perform this experiment. Once the researcher obtains a pure genomic sample, the first stage of exome sequencing begins: library preparation.
At this stage, the desired length of each genomic fragment is an essential consideration. As discussed before, human genes rarely exist as one continuous stretch of DNA, and instead are broken into gene encoding parts that are interspersed with non-coding DNA. Over 80% of all gene encoding sequences are less than 200 base pairs (bp) long.
As it’s only the exome (all of the exons) that the researcher wants, it’s very common to break the fragments into pieces ≤200 bp long to make sure unnecessary sequences are minimized in later stages of the experiment. Understandably the question of fragment length will always be application specific, and is a decision made based on appropriate and available sequencing technology. For the Illumina sequencing standard, which is commonly used in exome sequencing, fragments 100-200 bp long are made. To make these fragments, several commercially available methods exist on the market that simplify the process.
Next, the genome fragment soup needs to be prepared for sequencing. This is a 3-part process:
- Blunting: When an enzyme is used to chop the DNA into small parts, it leaves ends of uneven length on the double strand—one will have a few base pairs more than the other. Depending on which strand the overhang is on, the base pairs are either removed, or the missing base pairs are filled in by an enzyme producing “blunt” ends of equal length.
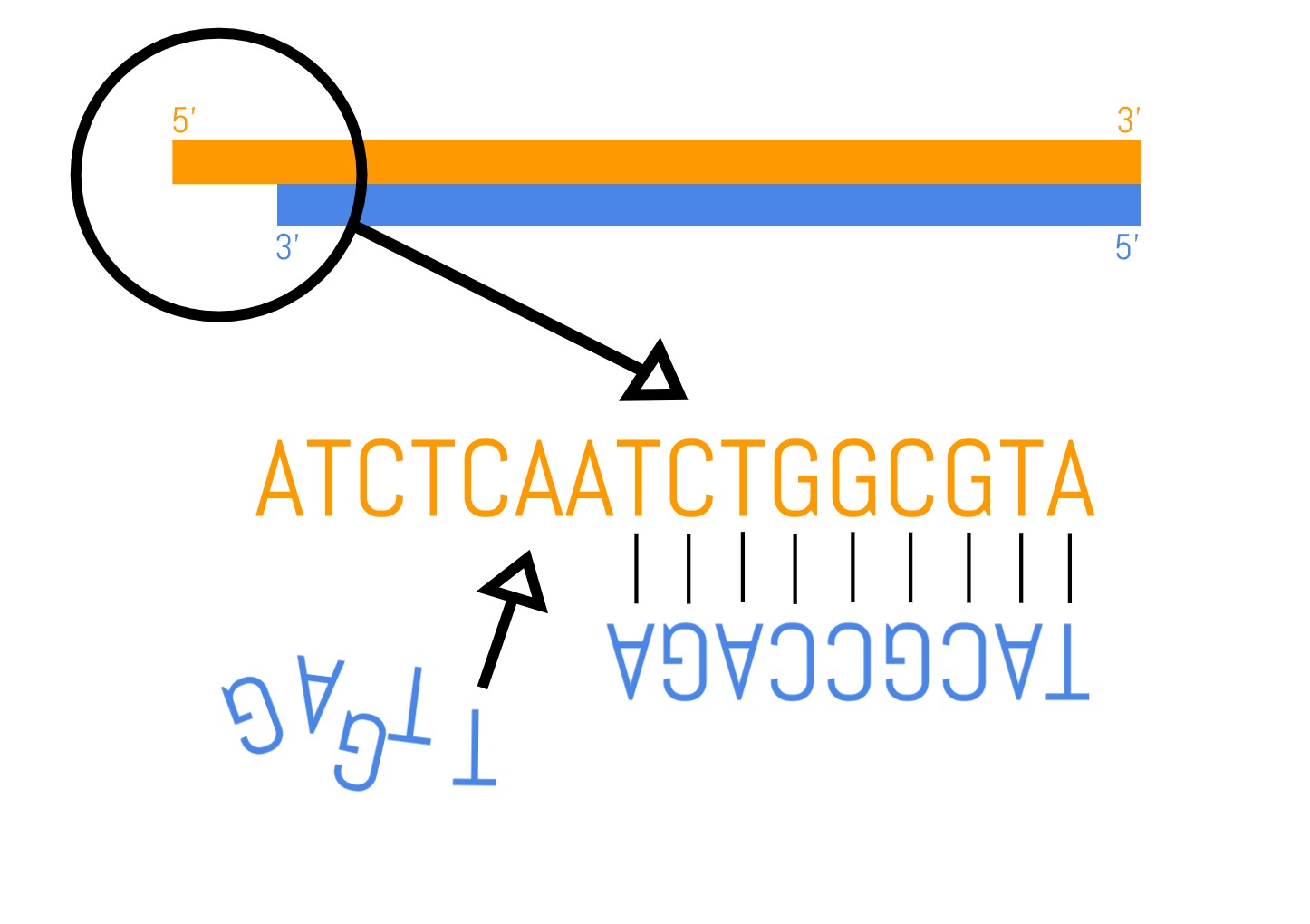
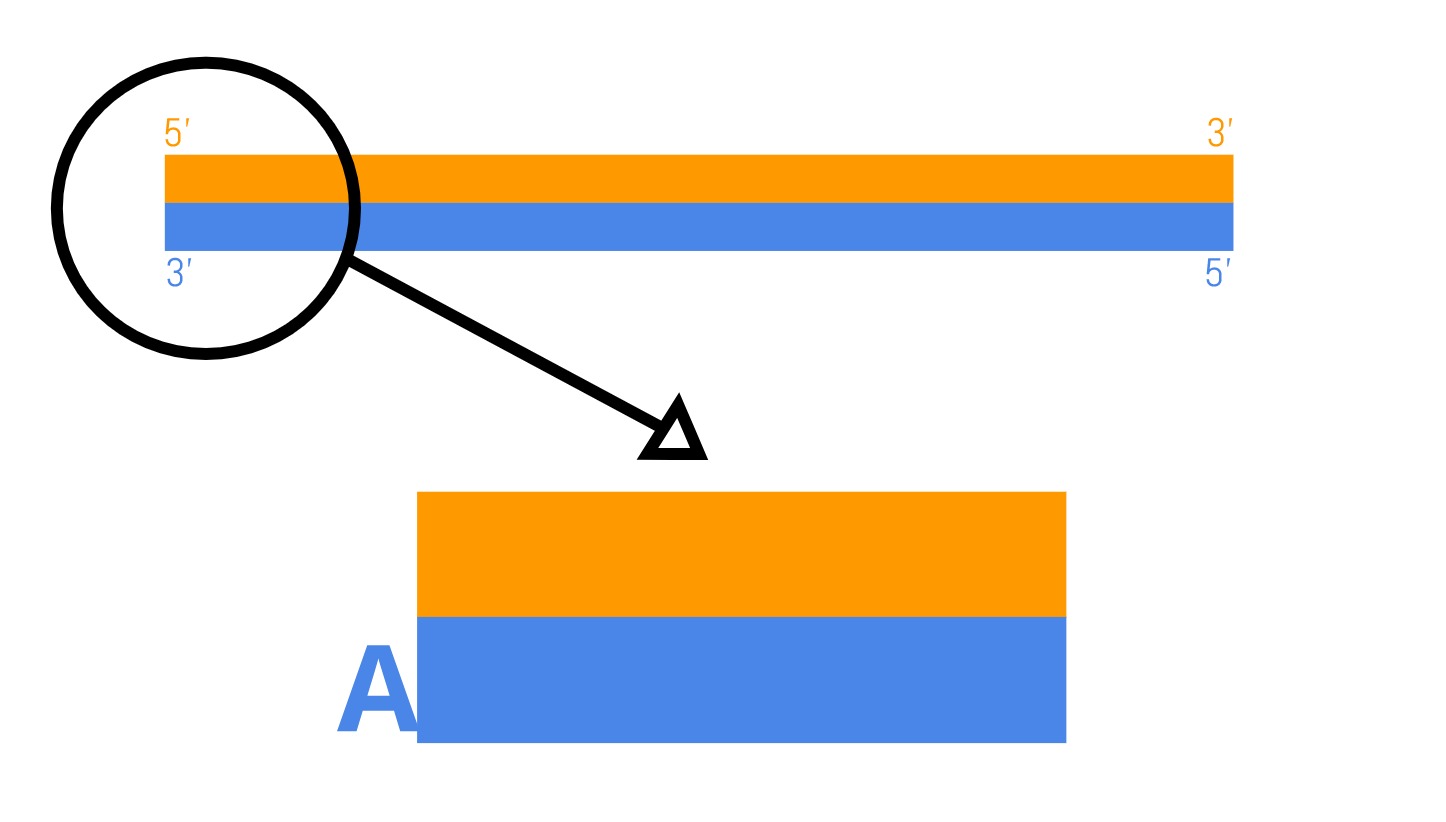
- A-tailing: The blunted ends are then modified by adding a single adenine (A) nucleotide that forms an overhanging “A-tail”.
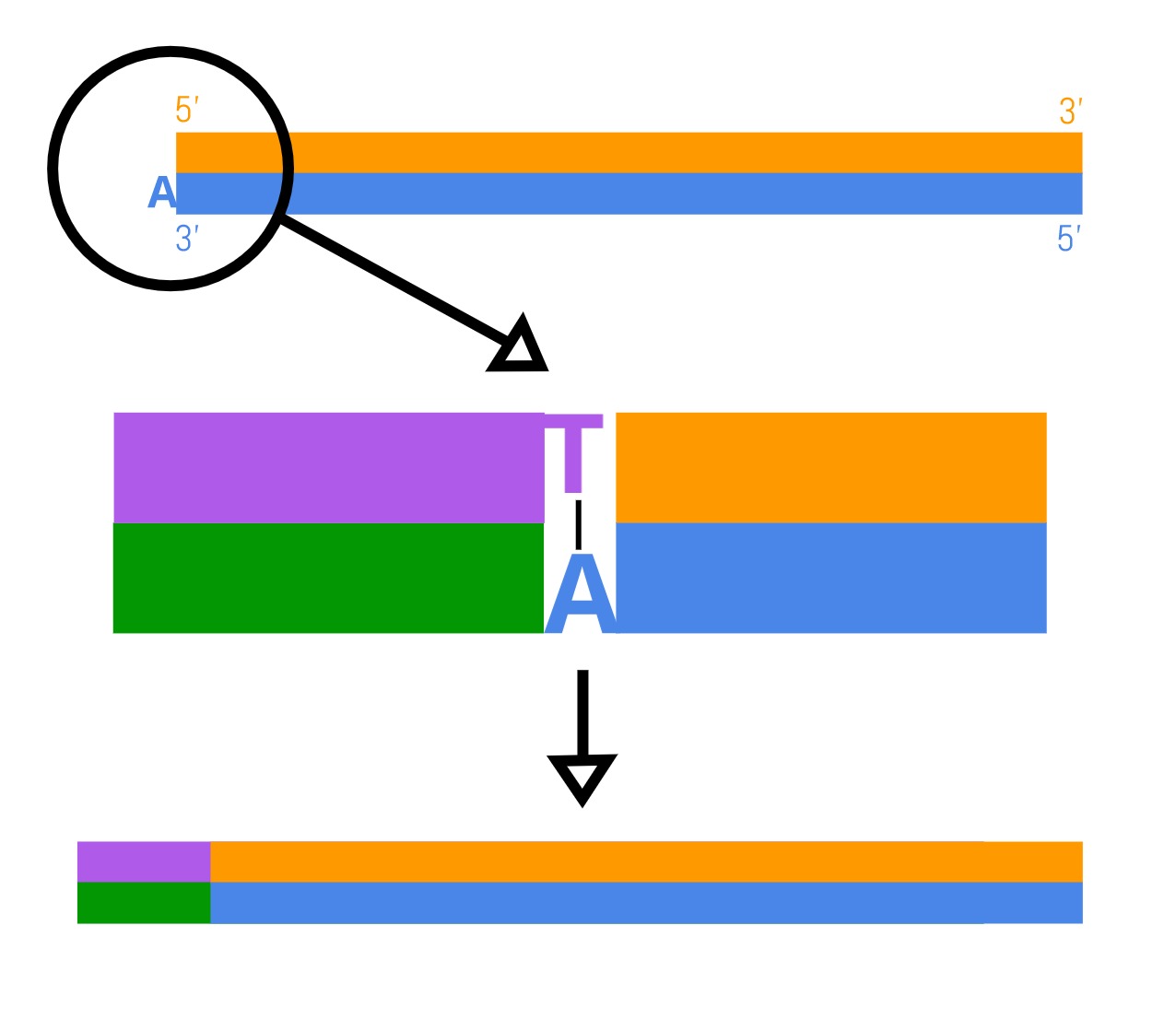
- Adapter ligation: Adapters are added to each sequence. High throughput sequencing requires specific DNA sequences called adapters that kick-start the DNA reading reaction. Where there is an overhanging A nucleotide, adapters with an overhanging thymine (T) nucleotide form base-pair interactions. An enzyme is used to stitch these two interacting fragments together using covalent bonds. Each genome fragment now possesses the same sequencing adapters and are ready to be sequenced.
One aspect of Twist Bioscience’s Human Core Exome Enrichment Kit, a complete kit for sequence library and human exome enrichment preparation, is a property called multiplexing. This allows the researcher to pool DNA samples from our exemplar patient with DNA samples from other patients who are undergoing the same screening. Their exomes can then be sequenced all together, only to separate each patient’s data post-hoc using computer programs.
Multiplexing is possible due to an extra DNA sequence added with the adapter sequence called a barcode. This is a known sequence that can be referenced in each sequencing read. There are many different barcodes available, meaning the researcher can treat Sample One with one barcoded adapter, and Sample Two with a different barcoded adapter. Multiplexing lowers both the cost of each sequencing reaction, and the time taken to complete the sequencing of multiple samples.
Once adapters are ligated onto each genomic fragment, the genomic library is prepared for the next stage of the exome sequencing protocol: Target Enrichment.
Here is the entire series:
Exome Sequencing 101: Part 1 – Library Preparation
Exome Sequencing 101: Part 2 – Target Enrichment
Exome Sequencing 101: Part 3 – Next-Generation Sequencing (NGS)
What did you think?
Like
12
Dislike
1
Love
3
Surprised
0
Interesting
1