Targeted RNA-Seq Expands the Toolkit for Early Cancer Detection

RNA-seq is an essential research tool that holds exciting prospects for the early detection of cancer. However, in clinical samples highly abundant mRNA and rRNA sequences obscure rarer RNA subtypes that may hold the keys to early cancer detection, such as long non-coding RNA. Here we dive into recent research from the University of Ghent outlining the use of targeted RNA-seq to enrich lncRNA from various sample types and discuss how recent advances make target enrichment an invaluable tool for high-resolution RNA-seq experiments.
As a rule of thumb, treating cancer is much easier and more successful when detected early. In a UK report from 2018, only 55% of cancers are detected at stage 1 or 2. Cancer type also plays a significant role in early detection. In the report, 90% of testicular cancer cases were detected early, compared to only 22% of pancreatic cancers (ref 1).
Researchers have long been searching for screening methods that are minimally invasive, non-damaging, yet sensitive enough to detect small populations of cancerous cells, aiding and improving early detection. Out of this effort has grown an increased excitement around a genetic element called long non-protein coding RNA (lncRNA).
lncRNAs belong to a diverse category encompassing any RNA molecule that is more than 200 nucleotides in length and does not translate protein-coding information. There are thousands of recognized lncRNA transcripts, some of which are believed to play an instrumental role in cancer development and progression. Cancer-associated lncRNA transcripts can be found circulating in a patient’s blood. This means that liquid biopsies utilizing RNA sequencing (RNA-seq) for the detection of lncRNA could be a valuable diagnostic tool.
Though, for the most part, we know very little about lncRNA transcripts, much less in what contexts they’re expressed. Their elusiveness is primarily due to technological limitations that make sequencing and identifying lncRNA very difficult (refs 2-4).
Fortunately, advances in oligonucleotide synthesis and RNA-seq are helping researchers unravel lncRNA and its many mysteries. Here, we’ll dive into a recent advancement in this field. Researchers from Ghent University, Belgium, have developed a highly sensitive RNA-seq method for identifying lncRNA transcripts from myriad sample types, providing a promising tool for early cancer detection.
Targeted RNA-Seq vs. Total RNA-Seq
Studying the sequence and function of lncRNA transcripts presents several challenges to researchers. Compared to messenger RNA (mRNA) and ribosomal RNA (rRNA), lncRNA is expressed in very low quantities (refs 3,5). This means that mRNA and rRNA obscure lowly expressed lncRNA transcripts during RNA-seq.
One way to overcome this challenge is through the use of targeted RNA-seq, wherein broad pools of genetic material are filtered to enrich specific RNA transcripts. Doing this allows sequencing resources to be focused on the selected transcripts and prevents masking of data from undesired, more abundant transcripts.
Target enrichment can be used for most RNA-seq projects where finer resolution data is needed, whether it’s whole exome sequencing or an analysis of the lncRNA transcriptome. In both cases, enrichment can be achieved by designing a panel of custom probes that are homologous to the target RNA transcripts.
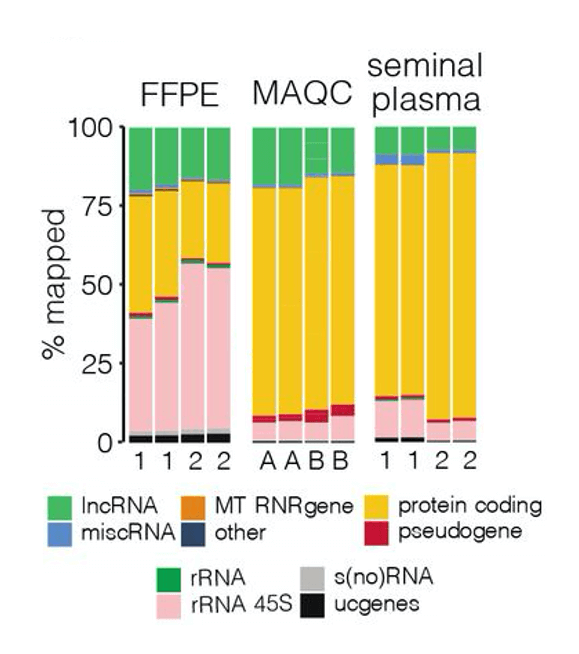
The advantages of a targeted RNA-seq approach are emphasized in a pre-print paper by Morlion et al. (2021). The team leveraged a custom panel of 565,878 target capture probes that were synthesized by Twist Bioscience. For comparison, the researchers also used total RNA-seq, which casts a wide net to detect many forms of RNA, including mRNA and lncRNA.
Using a variety of sample types cancer researchers may encounter, including seminal plasma, platelet-depleted blood plasma, formalin-fixed paraffin-embedded tumor samples (FFPE), and MAQCA/B (Quantitative PCR Human Reference Total RNA), the team found that targeted RNA-seq with custom RNA capture probes resulted in:
- 3.5, 4, and 8x increase in the percentage of reads mapped to lncRNA in FFPE, MAQCA/B, and seminal plasma samples, respectively
- 2x increase in uniquely detected lncRNA transcripts
- Reproducible identification of several thousand lncRNA transcripts that were missed by total RNA-seq
- Identification of five cancer-related lncRNA transcripts that were missed by total RNA-seq
- An average of 6x increase in coverage of cancer-related lncRNA transcripts
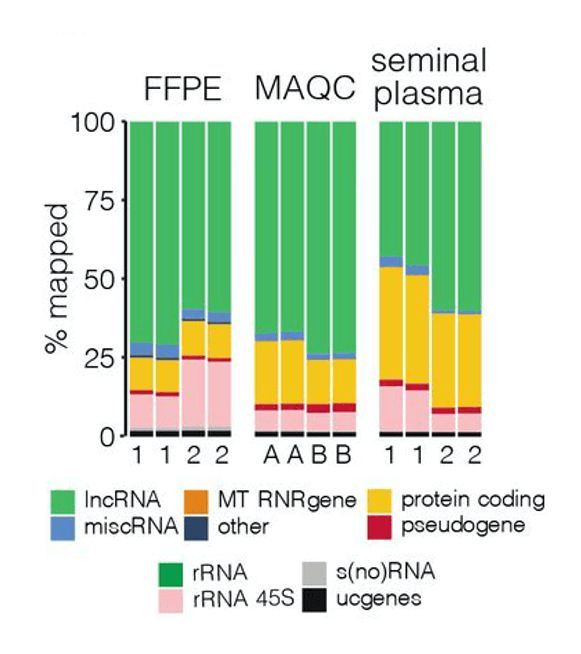
In short, by specifically designing a uniform library of probes to target and enrich for lncRNA transcripts, the team was able to greatly improve sequencing coverage, sensitivity, and reproducibility of sequencing relative to the current gold standard of total RNA sequencing.
Custom Target Enrichment Panels for Targeted RNA-Seq
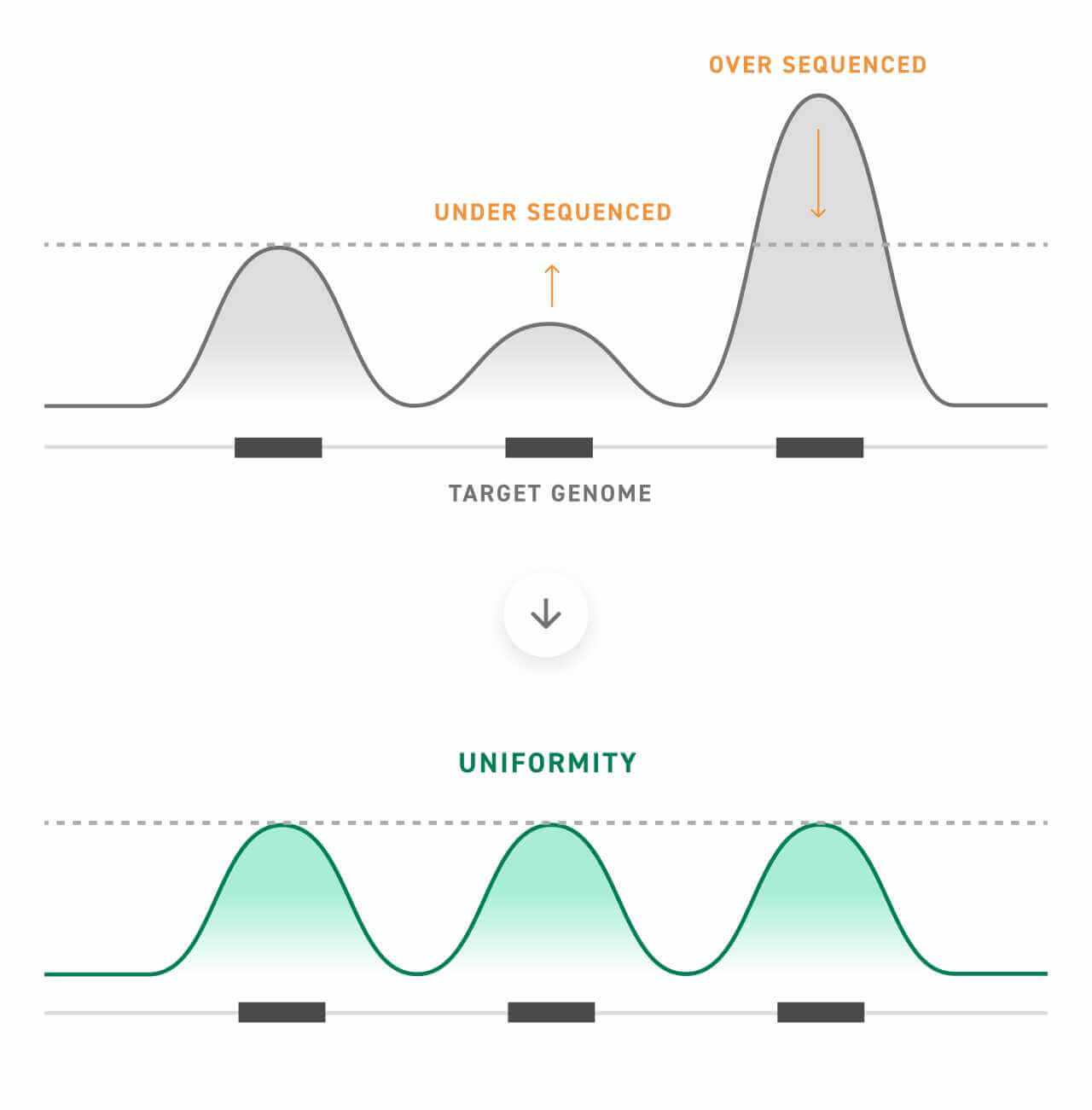
The findings presented by Morlion et al. contribute to the growing body of evidence suggesting that RNA-seq can become more sensitive when paired with target enrichment. However, realizing this potential requires the careful design of probe libraries optimized for both uniformity and a high on-target rate.
On-target rate is a measurement of how many of the captured sequences were intended targets, whereas uniformity is a measure of how evenly this capture occurs. Optimizing both uniformity and on-target rate improves sequencing metrics for a more sensitive experiment, especially important when working with an elusive nucleotide type like lncRNA.
At Twist, we specialize in this type of high-quality target enrichment panel design and construction.
To support this research, we delivered a custom panel of target enrichment probes. Leveraging our high-throughput silicon-based DNA synthesis platform allowed rapid production of a highly uniform panel of biotinylated, double-stranded DNA probes that were ready for experimentation.
Want to learn more about panel uniformity and target enrichment? Check out these resources:
- White paper: The Importance Of Coverage Uniformity Over On-Target Rate For Efficient Targeted NGS
- 15 minute Webinar: Custom enrichment for RNA sequencing of liquid biopsies
Clinical Significance of lncRNA
Mounting evidence suggests that lncRNA transcripts can serve several functions in basic cell biology, from transcriptional and post-transcriptional gene regulation to directing protein localization1. Some lncRNA transcripts also appear to be tumor suppressors while others are critically involved in cancer development and progression (refs 4,6).
Expressed in a highly cell-specific and time-dependent manner, lncRNA could be leveraged for both therapeutic development and diagnostics (ref 6). Here’s a good review on this topic.
More research is needed for lncRNAs to become clinically significant biomarkers. Researchers will need to be able to identify, quantify, and study the many different types of lncRNA that may be rare or lowly expressed under various conditions. To do that, they’ll need sensitive and efficient assays like the targeted RNA-seq assay described here.
Learn more about how custom target enrichment panels can support your research here >
References
- Public Health England. National Disease Registration Service: Case-mix adjusted percentage of cancers diagnosed at stages 1 and 2 in England, by Clinical Commissioning Group. May 2020. (Accessed 3rd June 2021).
- Yao, Run-Wen, et al. “Cellular Functions of Long Noncoding RNAs.” Nature Cell Biology, vol. 21, no. 5, 2019, pp. 542–551., doi:10.1038/s41556-019-0311-8.
- Bussotti, Giovanni et al. “Detecting and comparing non-coding RNAs in the high-throughput era.” International journal of molecular sciences vol. 14,8 15423-58. 24 Jul. 2013, doi:10.3390/ijms140815423
- Lorenzi, Lucía, et al. “Long Noncoding RNA Expression Profiling in Cancer: Challenges and Opportunities.” Genes, Chromosomes and Cancer, vol. 58, no. 4, 2019, pp. 191–199., doi:10.1002/gcc.22709.
- Djebali, Sarah et al. “Landscape of transcription in human cells.” Nature vol. 489,7414 (2012): 101-8. doi:10.1038/nature11233
- Shi, Ting et al. “Long Noncoding RNAs as Novel Biomarkers Have a Promising Future in Cancer Diagnostics.” Disease markers vol. 2016 (2016): 9085195. doi:10.1155/2016/9085195
What did you think?
Like
1
Dislike
0
Love
0
Surprised
0
Interesting
0