DNA is the unseen constant in our world. Not only do its As, Ts, Cs, and Gs form the basis of all life on earth, but DNA that’s shed from living things will pervade the environment around us and, in some cases, will long outlive us.
As a biomaterial, DNA is remarkably stable. When stored in a cool, dry climate—such as the deep interior of a cave, nestled within mineralized bone—DNA can be stable for hundreds of thousands of years1. It’s not surprising then that DNA can be found in the excavated remains of ancient human beings, presenting a window into the past through which researchers can study our evolutionary history.
The promise of ancient DNA lay in its information-carrying capacity. Patterns stored in DNA can tell us about the lives of ancient hominids, the population dynamics of their time, and, potentially, how modern humans came to be. But, extracting this information from such weathered samples is no small task. Not only is it technically challenging, but the process of extracting DNA is destructive, meaning that these precious relics must be damaged, if not outright destroyed. Therefore there is little tolerance for error when working with ancient DNA—even small inefficiencies can cause irreparable loss. With such high stakes, it’s critical that researchers have access to the best tools for the job.
Tools for peering into the past
In 1984, researchers at the University of California Berkeley used molecular cloning techniques to produce the world’s first ancient DNA sequence: two short fragments (<300bp) of mitochondrial DNA wrenched from the preserved muscle tissue of a 140-year-old quagga specimen (Equus quagga)2. While shedding interesting light on the evolutionary history of equines, the study proved foundational as a proof of concept that even time-worn DNA could provide valuable information about the past, thus establishing the new field of paleogenomics.
Molecular cloning may have enabled our first foray into paleogenomics, but it quickly fell to the wayside as more advanced sequencing technologies were adopted 3,4. Now, like a surgeon reaching for a scalpel, researchers studying ancient DNA will often reach for target enrichment panels upstream of next-generation sequencing (NGS) to help them extract reliable data from ancient samples.
In contrast to the limited nature of earlier technologies (like molecular cloning and PCR), NGS enables massively parallel sequencing and broad coverage, such that an individual’s entire genome can be sequenced in a single run. The first use of NGS technology for ancient DNA analysis came in 2006 with the sequencing of ~13 Mb of DNA extracted from a 28,000-year-old Wooly Mammoth and ~1Mb of DNA from a Neanderthal4. The massive gains in sequencing information provided by NGS catalyzed a boom in paleogenomics (see Fig. 1)).
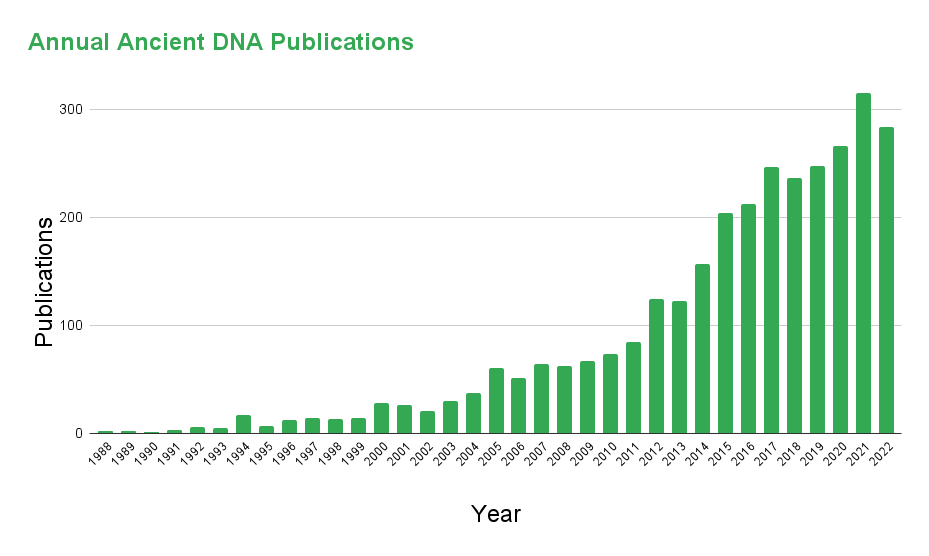
Yet, for all its benefits, NGS can be an extremely costly process, particularly with respect to paleogenomic studies. Typically, ancient DNA represents less than 10% of the total DNA found among unearthed remains, with the remaining 90% (and often much more) consisting of exogenous DNA shed from bacteria, fungi, or modern humans 4. If NGS technology is applied without prejudice, considerable resources will be spent sequencing this uninformative, contaminating DNA. This is where target-enriched NGS (hereafter referred to simply as targeted NGS) is beneficial.
Targeted NGS panels in Paleogenomics
Targeted NGS allows researchers to focus their sequencing resources on just the genetic fragments that are of interest to their study. This is typically done through the synthesis of biotinylated oligonucleotide probes that are designed to be complementary to a specific target sequence. When incubated with the sample DNA, these oligonucleotide probes will bind to their complementary target sequence. Addition of streptavidin-coated beads then allows for the probes—and any sequences they’re bound to—to be immobilized. The remaining DNA in the sample can be washed away, leaving behind only the bound fragments of interest.
By removing the excess DNA, researchers are able to concentrate their sequencing resources on specific genomic targets and, in doing so, gain deeper sequencing coverage. This often translates to more accurate sequencing results with less capital investment (see our recent blog about the benefits of target enrichment to learn more).
To appreciate the powerful potential of target enrichment in studying aDNA, consider some back-of-the-envelope math. To perform whole genome sequencing of a modern human genome at the standard 30x coverage would necessitate about 400 million sequencing reads. However, ancient DNA samples are often contaminated with myriad non-human DNA. And, the ancient human DNA that is present is often degraded, such that less than 1% of sequences produce usable data. To achieve the necessary coverage over variants of interest using a broad, non-targeted strategy, one would need to push the total sequencing reads up into the billions, quickly making the experiment prohibitively costly and impractical. With enrichment, we can target just the variant regions of interest while also removing much of the contaminating DNA sequences. As a result, researchers can extract valuable information from many hundreds of thousands of variants in ancient samples with as little as 25 million reads per sample.
Since 2015, the paleogenomics field has largely relied on a specific target enrichment panel known as the “1240k reagent,” which is a collection of oligonucleotide probes that are designed to capture DNA fragments containing any of 1.24 million polymorphisms that are considered informative. As of October 2022, this panel had been used to generate >70% of the world’s genome-wide ancient human DNA data 5.
Though it is a widely used tool, there are limitations to the 1240k reagent, including variability in enrichment effectiveness, bias in allele capture, and—due to the absence of a commercial product—the high cost of synthesizing the target probe sets 5. Collectively, these limitations both make it difficult to compare data sets within the field and prevent all but a few laboratories from engaging in genome-wide studies of ancient DNA. To reduce these issues, leading researchers have worked to improve the design and accessibility of ancient DNA target enrichment panels.
🧬 Types of DNA Decay
There are many ways for DNA to degrade over time. Here are just a few:
- Fragmentation: The chemical bond between the sugar-phosphate backbone and the purine base is susceptible to hydrolysis which results in an abasic site in the DNA strand. An abasic site can, in turn, result in cleavage of the strand. This is why ancient DNA typically results in short fragments when extracted.
- Deamination: Another natural form of DNA decay is the deamination of cytosines to a uracil base. This is an important change because it will cause a polymerase to incorporate a base substitution during replication, resulting in both a C to T, and a G to A substitution. Both of these could confound the sequence analysis.
- Lesions: Many different modifications can occur to DNA which can result in cross-linking. This modification can block enzymes from performing their functions and prevent copying of the DNA strand.
Learn how Twist is helping researchers work around DNA decay and tap into our ancient past in this webinar: Using Twist technology to efficiently enrich 1.4 million SNP targets for ancient DNA analysis
The Twist Ancient DNA Panel
In 2021, Twist Bioscience made a commercially available target enrichment panel for aDNA that enriches for the same 1.24 million polymorphisms that are covered by the 1240k reagent, plus additional content of interest. Nadin Rohland, Swapan Mallick, Matthew Mah, Robert Maier, Nick Patterson, and David Reich at Harvard University set out to perform an independent comparison of an optimized Twist Ancient DNA Panel that they designed with both the 1240k reagent and another commercially available panel5. Results showed that Twist’s panel provided highly uniform coverage with less bias toward fragment length, 1.2 to 1.4x increase in overall SNP coverage, and the ability to provide such high-quality data that it can be co-analyzed with shotgun sequencing data without substantial bias (a longstanding, yet unmet need within the field).
The high performance of the Twist Ancient DNA Panel is such that Harvard University lab has since transitioned away from the use of the 1240k reagent, favoring instead the newer, optimized panel, using the Twist reagent already more than 10,000 times5,6. As a commercially available resource, too, Twist’s panel helps eliminate the economic burden of synthesizing target enrichment probes and opens the door to wider participation in paleogenomics research.
References
- Allentoft, Morten E., et al. “The Half-Life of DNA in Bone: Measuring Decay Kinetics in 158 Dated Fossils.” Proceedings of the Royal Society B: Biological Sciences, vol. 279, no. 1748, 10 Oct. 2012, pp. 4724–4733, https://doi.org/10.1098/rspb.2012.1745.
- Higuchi, Russell, et al. “DNA Sequences from the Quagga, an Extinct Member of the Horse Family.” Nature, vol. 312, no. 5991, Nov. 1984, pp. 282–284, ui.adsabs.harvard.edu/abs/1984Natur.312..282H, https://doi.org/10.1038/312282a0.
- Danielewski, Mikołaj, et al. “Methodological Changes in the Field of Paleogenetics.” Genes, vol. 14, no. 1, 16 Jan. 2023, p. 234, https://doi.org/10.3390/genes14010234.
- Shapiro, B., and M. Hofreiter. “A Paleogenomic Perspective on Evolution and Gene Function: New Insights from Ancient DNA.” Science, vol. 343, no. 6169, 23 Jan. 2014, pp. 1236573–1236573, science.sciencemag.org/content/343/6169/1236573.full, https://doi.org/10.1126/science.1236573.
- Rohland, Nadin, et al. “Three Assays for In-Solution Enrichment of Ancient Human DNA at More than a Million SNPs.” Genome Research, vol. 32, no. 11-12, 1 Nov. 2022, pp. 2068–2078, https://doi.org/10.1101/gr.276728.122.
- Liu, Yue-Chen, et al. “Ancient DNA Reveals Five Streams of Migration into Micronesia and Matrilocality in Early Pacific Seafarers.” Science, vol. 377, no. 6601, July 2022, pp. 72–79, https://doi.org/10.1126/science.abm6536.










