iGEM 2016 – Giant Jamboree Wrap Up

The rules are simple. Each team is shipped the same box of modular genetic parts in the Biobrick DNA format. Combining these with their own innovations, teams develop a project that advances the synthetic biology field.
Teams use the genetic parts to design projects that span a wide range of research areas. Projects this year included introducing new, useful functions into organisms, developing tools to improve the genetic engineering toolbox for a particular organism, modeling interesting biological processes to improve engineering processes, designing new software or hardware to enhance synthetic biology, or expanding the scope of an already existing technologies within the field. Teams then come together to show off their hard work at the Giant jamboree – a huge celebration of everybody’s achievements held in Boston MA.
While all of the projects can be deemed a success, iGEM is after all a competition, and prizes were awarded to the top performing teams. The Grand Prize winners for the High School, Undergrad, and Overgrad levels all made incredible advancements to the synthetic biology field, with projects that rival those from the world’s top researchers.
Here are the iGEM winners for 2016:

HSiTAIWAN found that the public is largely unaware of the potential danger, it is medicine after all! As a result, the government treats possible issues with Chinese medicine as a low priority. Also, with many herbs being traded across Asian boarders, it is difficult and expensive to test the quality and contamination of all of the herbs used in Chinese medicine.
This is an issue with huge socioeconomic importance—if it gets out of hand, the practice of traditional medicine could disappear, erasing a part of Asian culture that has been around since before recorded history.

Their sensors work by expressing proteins that sensitively and selectively bind the contaminants, driving expression of a reporter gene. For example, one of their circuits expresses the lead-binding protein PbrR. When bound to lead, PbrR then binds to the pbr promoter to activate transcription. Their circuit contained a Green Fluorescent Protein (GFP) expression cassette under the control of the pbr promoter, and therefore GFP levels could be measured to determine lead levels in a sample.
Imperial College: Ecolibrium (Undergrad)

Finding a set of growth conditions that equally balances the two culture’s fitness, allowing both to survive, is a tightrope walk. Even the slightest condition change may favor one species over another.
To solve this problem, Imperial’s Ecolibrium used a technology called Small Transcriptional Activating RNA (STAR), an RNA-based switch. STAR was introduced into both co-culture species, engineered to turn off the bacteria’s genes that control cell division in the presence of a chemical “on” signal. The bacteria were then engineered to always make this “on” signal, essentially giving them 2 choices: kick out the now harmful machinery, or find a way to switch off STAR.
Here’s where things get really clever! Imperial provided each species with a second RNA technology called anti-STAR which is activated by a second chemical “on” signal. However, this time the on signal is only made by the other bacteria. This way, for either species to survive, they have to work together.
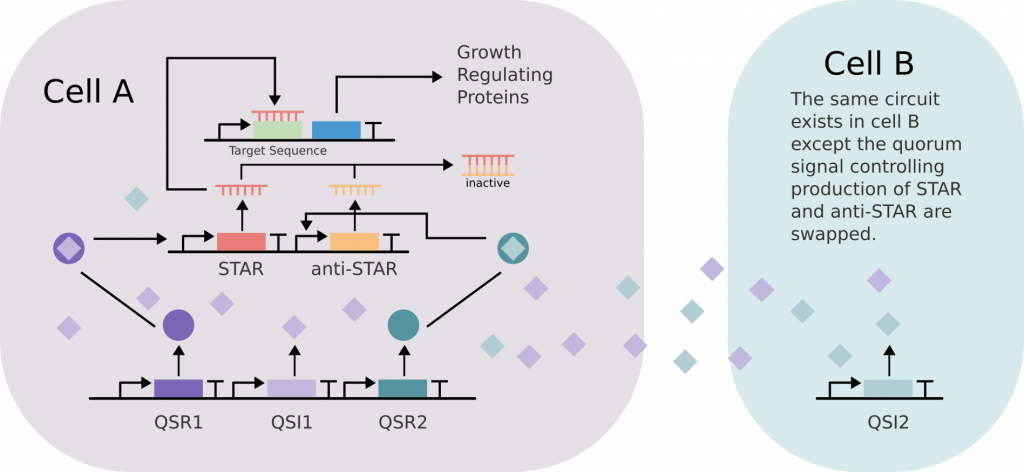
Now, both population densities are intrinsically linked. The Imperial team can finely control the population dynamics of the co-culture by changing the strengths of the promoters controlling the production of STAR, anti-STAR, and their corresponding signaling chemicals.
As a final impressive addition, Imperial also used computer models to fully predict how the population dynamics of their co-culture system will change over time given certain conditions. These models were integrated into their software called ALICE that allows researchers to precisely design their own co-culture experiments.
The Imperial iGEM team envisioned new applications for precisely controlled co-cultures. These are very well described on the project description page of their wiki, but two of our favorites were:
- Using a co-culture to model financial dynamics: Fine control over population density in bacterial co-cultures could model co-dependent real-world systems like financial banks. At its simplest level, when a stress is introduced that affects only a single organism (a single bank), the widespread effects on the other organisms (banks) can be observed.
- Biosynthesis of complex materials: Many biological systems — the iridescence of a butterfly wing, for example — gain their properties at the microscopic level. That makes these properties difficult to replicate in materials production. Motile, communicative co-cultures could be engineered to synthesize materials by forming complex, microscopic population patterns. Commodity material properties could then be engineered with precise intricacy.

Organ transplantation is hailed as a modern medical marvel, saving hundreds of thousands of lives over the past century. Over 2,000 heart transplants are performed in the US every year. Yet a predicted 800,000 people have symptoms of severe heart failure, and are in desperate need for transplantation to improve both quality of life and life expectancy. Long waiting lists are typically caused by a limited pool of viable organ donors.
The Biotink project addressed this problem by bringing a new solution for the assembly of stem cell-derived artificial tissues to the table—3D printing.
An effective alternative would involve controlling the differentiation of patients’ own stem cells so there would be no risk of organ rejection – these cells would then be used as ink in a 3D printer, that doesn’t print plastic but instead prints whole organs! Many organs are complex networks of various cell types patterned at the microscopic level, and fine details like capillary networks are not possible to print with the resolution of current 3D bioprinting technology.
Their system centers around a strong binding interaction between the biological molecule biotin and the protein streptavidin. By engineering synthetic fusions of protein parts, cells can be made to display either a biotin molecule or streptavidin variants on their surface.
Cell aggregation is controlled by the addition of engineered “linker” molecules on the printed cells. Using different combinations of linkers and cell surface display molecules aggregation could be regulated at the cellular level. The Munich team developed a number of biotin- or streptavidin-based linker molecules that behave in a predictable way. Depending on the linkers each cell type is expressing on its surface, various predictable aggregation patterns can be programmed (see image).
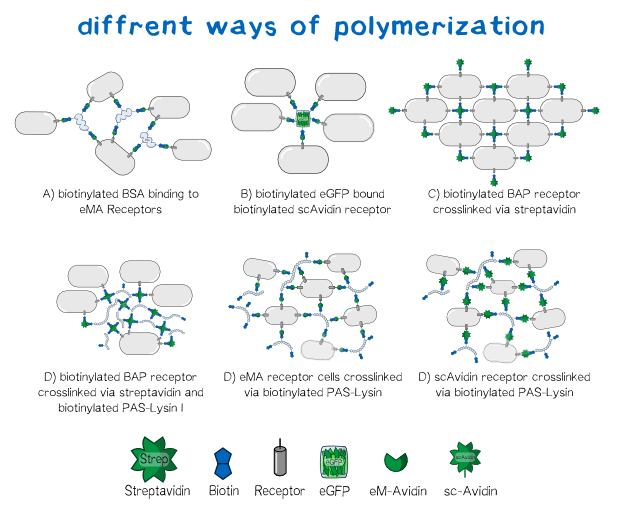
Armed with this toolbox, and some careful design, a large array of microscopic structures could be formed depending on the bio-printing requirements. The Munich team went on to verify the expression, localization, and viability of their surface binding proteins, develop, express, and characterize their linkers, and show early evidence that cell aggregation could be tightly controlled. They were then able to build the hardware needed for a 3D printer, and begin bioprinting.
This just scratches the surface of what the LMU-TUM Munich team achieved – our post really doesn’t do the scale of their project justice. Check out their wiki to see the immensity of it all! Additional amazing achievements (do not exhaustively) include:
- Characterization of the enzyme biotinidase for the degradation of the biotin-streptavidin-cell mesh
- Chemical engineering of different biotin molecules allowing for controlled biotinidase-based degradation
- Early stage development of machinery to aid capillary formation
- Design and early stage construction of an electrical-biological hybrid device that can be transplanted into patients to deliver targeted therapeutics
- A biotin-streptavidin-based kill mechanism to destroy cells in the therapeutic, if necessary
- Converted a 3D printer to a bioprinter by 3D printing the bioprinter parts, and made the instructions available for anyone to use
- Developed new software to control their 3D printer
- 3D printed their logo as an aggregating tissue!
- Genspace: Droughtbusters – A community lab that characterized proteins from the tardigrade — a little organism that is able to survive desiccation — and showed that desiccation-protecting protecting proteins from tardigrades can also protect E. coli.
- NCTU-Formosa: Pantide – Ideal insecticides will kill insects, but be harmless to everything else. Pantide is a peptide-based insecticide technology inspired by spider venom.
- Sydney Australia: FRES(H) – Did you know over half of all fruit is wasted before it even gets into your fruit bowl? FRES(H) is an ethylene biosensor that can help mitigate this waste.
- Groningen: CryptoGE®M & Edinburgh UG: BABBLED – Digital data storage comes in two forms, short-term and long-term. Short-term data storage is what your computer uses. Long-term storage involves storing data on tapes in huge archives that have to be regularly replaced. Both teams worked on projects very close to our hearts atTwist Bioscience, developing new ways for encoding digital data for long-term storage using DNA.
- Tec-Monterrey: Metal Recovery from E-waste – Used electronics create a lot of waste, they sometimes contain toxic chemicals, and they are often difficult to recycle. Tec-Monterrey developed a bacterial system that could help, using bacteria to remediate useful metals from E-waste, while also degrading harmful chemicals like cyanide.
A huge congratulations goes out from us at Twist Bioscience to all involved in iGEM this year—it has been yet another outstanding competition and show of community spirit. We can’t wait to see what next year has in store!
What did you think?
Like
Dislike
Love
Surprised
Interesting