Looking for information on the current outbreak of mpox virus? Check out our recent article on Synthetic DNA’s Critical Role in Infectious Disease Research.
On September 20, 2017, the World Health Organization (WHO) learned that an outbreak of mpox had hit Nigeria. This was an outbreak unlike any the country had ever seen. Related to smallpox, mpox (formally known as Monkeypox) is a rare disease caused by the mpox virus. The virus first infects people through contact with wild animals and then spreads from person to person. Like many other poxviruses, the infection causes painful lesions on a patient’s skin. In the two months that followed the initial reported case, 172 suspected and 61 confirmed cases were reported across the country, including one fatality. Until now, mpox had been extremely rare in Nigeria, with the last reported case occurring in 1978.
In Africa, incidences of mpox have been linked to two distinct clades, distinguished by their origin in the Congo Basin or Western Africa. Outbreaks associated with the Western Africa clade have been found to be considerably less virulent than outbreaks associated with the Congo Basin clade.
With this in mind, the U.S. Army Medical Research Institute of Infectious Diseases (USAMRIID), in partnership with the Institute Pasteur in Dakar, Senegal (IPD), and the Nigerian Centre for Disease Control, sought to determine the origin of the recent outbreak of Nigerian mpox to better understand its potential impact to public health. Using a genomic capability recently established at USAMRIID at the IPD, the research team sequenced the virus’s genome from blood samples collected from the infected patients.
However, sequencing viral genomes directly from blood samples is challenging, due to low levels of virus present in the sample - almost all of the genetic material present is DNA and RNA from the patient. Identifying the viral sequences amongst the human sequences is a “needle in a haystack” problem. In order to increase sensitivity for the viral component in the sample, a target enriched next-generation sequencing (NGS) approach was taken. The study makes use of a pan-viral panel, designed by Illumina in collaboration with USAMRIID and synthesized by Twist Bioscience. The panel enables researchers to enrich for viral targets known to infect humans, an effort that was reported in a paper recently published in The Lancet. We take a closer look into the results of this study here.
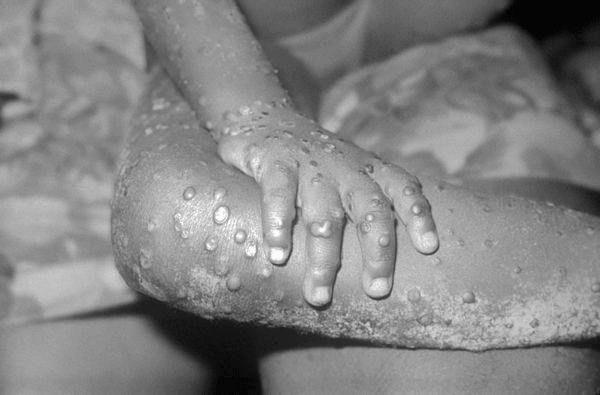
Lesions on the skin of a child suffering from mpox (Source: Centers for Disease Control and Prevention)
Where does Mpox come from?
Mpox gets its name from its first discovery in macaques (a genus of monkey) in 1958. However, despite its name, small rodents are believed to be the main carriers of the virus, maintaining the virus in nature until spillover events into humans and other mammals occur. The first human mpox case was identified in 1970 in the Democratic Republic of Congo. Mpox can cause symptoms and fatal illness that present as a milder version of the related human smallpox which was eradicated in 1980. However, it is thought that the smallpox vaccine being no longer required has led to the increase in human mpox cases, as the vaccine was also highly effective at protecting against the mpox virus.
In recent years, mpox has been prevalent in Central Africa. The most recent outbreak occurred in the Central African Republic in 2016, with 27 reported cases. Between 2009 and 2014, the Congo Basin (Congo and the Democratic Republic of Congo) also saw 333 cases. However, Western Africa hadn’t seen mpox in over 40 years. In the past, mpox patients were infected with one of two mpox virus clades: central and western. Historically, the viral strains from the clade affecting patients from Central Africa manifested a far worse disease progression than the strains from Western Africa. Before the current outbreak, there were no reported mpox-related fatalities from western cases, with an estimated mortality rate of around 1 percent while cases from Central Africa have seen mortality rates as high as around 20 percent (see image below).
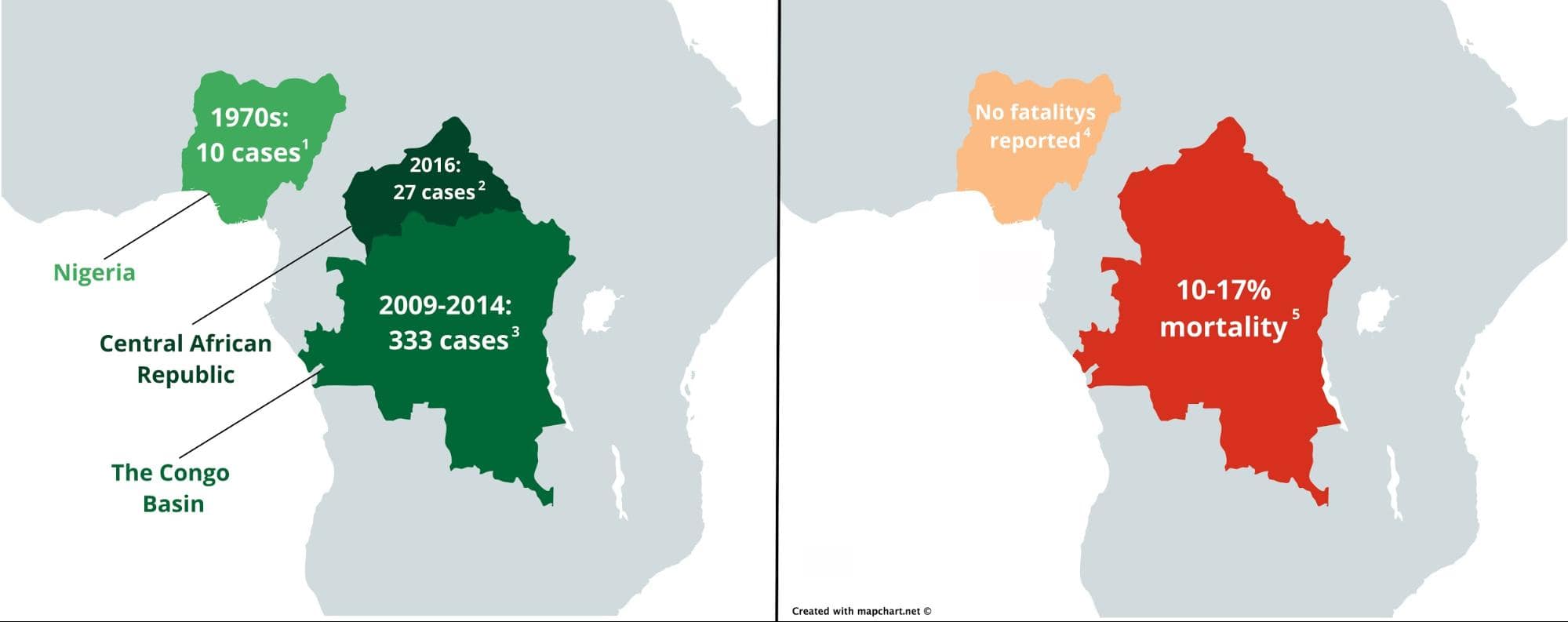
Outbreaks of human mpox virus reported before the current outbreak (left) and their mortality rates (right) in Africa [refs 1-5]. Maps were created with Mapchart under the creative commons license and edited in Gravit Designer.
When spillover events of mpox virus occur, it has often been linked to poor sanitation, and improper handling of patients caused the disease to spread. However, the recent cases of mpox in Nigeria are unusual. Their geographical occurrence is sporadic with little clustering, and only 7 percent of the cases in Africa are linked to human-human spread.
Increased prevalence and severity compared to previous Nigerian epidemics has made it possible for the Central African strain of smallpox to spread to Nigeria. A better understanding of the strain of mpox causing the Nigerian epidemic is imperative to understand the potential fallout, spread, and impact the disease could have on the Nigerian population, allowing world health agencies to better support Nigeria with the outbreak.
Tracking a virus
USAMRIID’s mission is to provide leading edge medical capabilities to deter and defend against current and emerging biological threat agents.
Since its inception in 1969, USAMRIID has spearheaded research to develop medical solutions—vaccines, drugs, diagnostics, and information—to protect U.S. service members from biological threats. While the Institute’s primary focus is on protecting military personnel, its research has applications that benefit society as a whole. USAMRIID also plays a key role in the investigation of emerging diseases throughout the world.
USAMRIID microbiologist Michael Wiley, PhD, helped coordinate a recent study in collaboration with the IPD in Senegal using next-generation sequencing of patient samples to determine whether they were carrying genetic material from the mpox virus. With the discovery of viral genetic information, comparing similarities and disparities in sample sequences allows Wiley’s team to draw links between existing strains and pinpoint the origin of the outbreak.
However, sequencing and extracting information about mpox viruses isn’t straightforward. Wiley’s team only had access to patient blood samples. Typically, researchers have two options for identifying viral sequences from blood samples:
Use DNA or RNAseq, next-generation sequencing methods, to sequence all nucleic acids in a patient sample to identify the presence of viral sequences.
Use specially designed primers that specifically target viral DNA/RNA, allowing for amplification of viral genetic material with PCR for increased resolution during next-generation sequencing.
There are pitfalls to both methods. In an infected patient blood sample, there is a small proportion of viral-origin genetic material compared to the majority genetic material which belongs to the host. In a PCR-based approach, some hypothesis of the infectious agent strain is needed, as reliable PCR requires amplification primers that are perfect matches to the viral DNA/RNA sample. In a scenario in which there are multiple potential viral agents, or if full characterization of the virus’s genetic makeup is required, multiple PCR assays need to be carried out to accurately identify the right virus and strain. Sequencing the transcriptome also has its drawbacks. This approach typically requires a lot of sequencing, making it not only a costly exercise but also one that will require long analysis times. Both screens are often slow, leading to a delay in obtaining actionable answers. Instead, Dr. Wiley sought a more efficient method of viral characterization.
Target enrichment uses DNA-based hybridization probes to physically isolate specific sequences of interest out of a mixed genomic sample. This method enables researchers in the field to isolate all the information needed about the disease from a single experiment from a complex source like a blood sample.
Development of a Pan-Viral Target Enrichment Panel occurred as a collaborative effort between the USAMRIID, Illumina, and Twist Bioscience. This panel consists of probes complementary in sequence to viral sequences that have been associated in human disease. With this panel, viral sequences can be enriched for and sequenced at high resolution using next-generation sequencing. This leads to high sensitivity detection and confident identification of viral sequences, even at low copy numbers.
The probe set contains over 700,000 probes that target over 1,000 viral species including mpox, compiled from the RefSeq database. By using this probe set, even tiny concentrations of viral genetic material can be separated from a mixed sample and amplified. Therefore, the sequencing experiment only focuses on the genetic material of interest, regardless of the sample origin and condition.
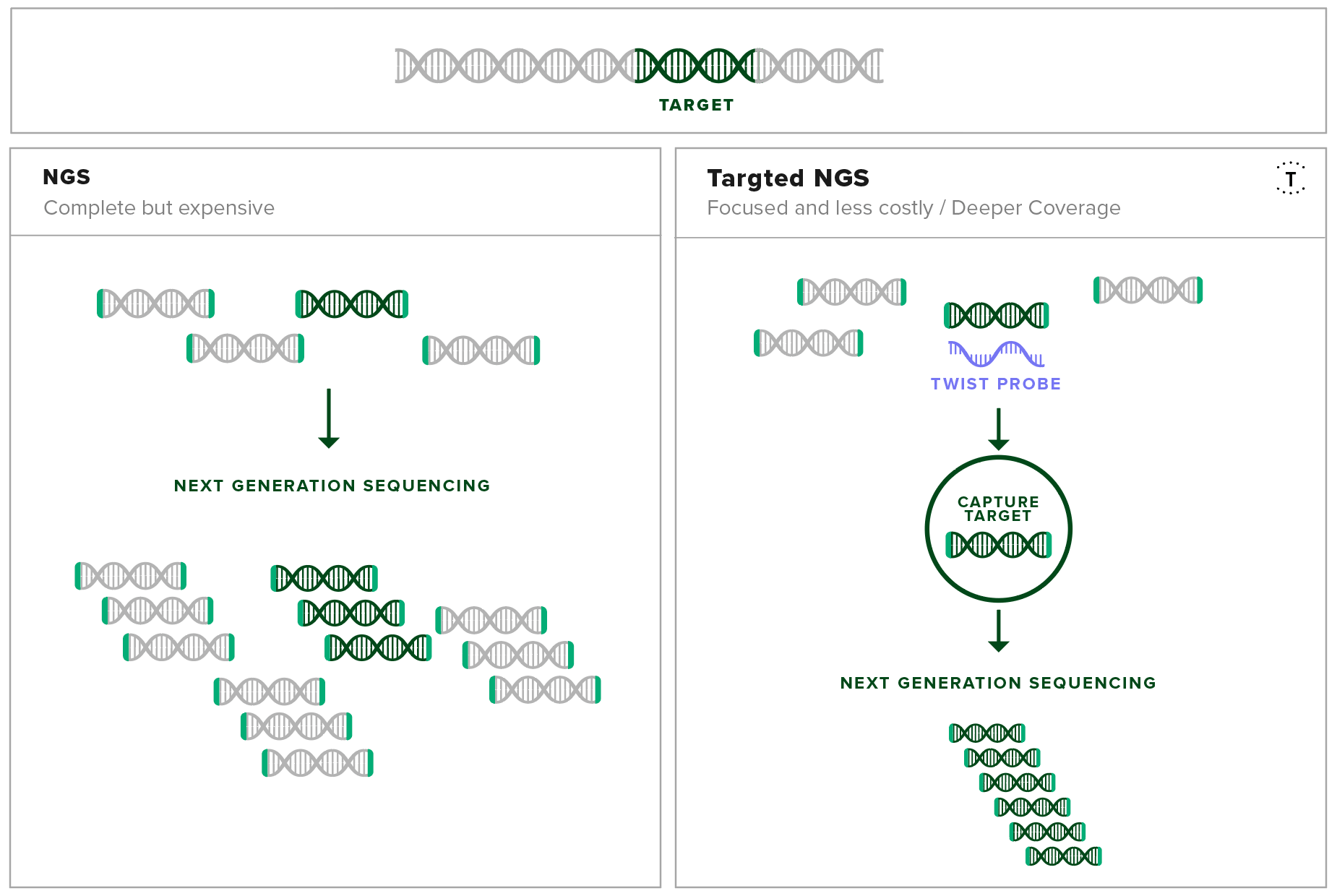
Target capture increases the resolution of the important fragments in next-generation sequencing experiments
By screening with the pan-viral target enrichment panel, the researchers in Senegal identified 22 out of 29 blood samples as positive for mpox. When these sequences were compared to the pre-existing data available on the genetic makeup of mpox, they were identified as being closely related to other mpox viruses of West African origin. This gives strong evidence that the current outbreak of mpox hasn’t spread out of Central Africa, and instead is more likely to be a “spillover event” from animal sources.
Zoonotic spillovers are defined as events where pathogens jump from their typical animal host to a human host. Spillovers occur from a compound of factors:
Viral incubation in the animal host causes the accumulation of mutations that allow the virus to survive in the new host.
Continued or increased release of pathogen particles from hosts or increased exposure to animal hosts containing the viruses increase the chance of a jump.
Infrastructure, sanitation and healthcare access issues increase the chance of animal host contact, decrease the effects of herd immunity from vaccination, and increase the chance of an infection event occurring.
When asked about the benefits of target capture, Dr. Wiley said, “Sensitivity is a big problem when screening for viruses in biological samples. Without an approach like the Pan-Viral Target Enrichment Panel, these experiments would have taken much longer and many more iterations, and would have required more resources to get the same results.”
Armed with the Pan-Viral Panel, researchers in Senegal were able to effectively screen blood samples for viral content with high precision, and provide rapid answers about the genetic makeup and origin of the mpox virus epidemic that has hit Nigeria recently. In the future, researchers are optimistic that the Pan-Viral Panel will allow a more simplified way of tracking emerging viral infections by enabling a more sensitive and streamlined method of screening patient samples, aiding in preparation and decision making to lessen the impact of disease epidemics on a global scale.
You can learn more about how Twist Bioscience supports target enrichment experiments here.
References:
- Faye, O., Pratt, C.B., Faye, M., Fall, G., Chitty, J.A., et al., 2018. Genomic characterisation of human Monkeypox virus in Nigeria. The Lancet Infectious Diseases 18, 246. https://doi.org/10.1016/S1473-3099(18)30043-4
- Brown, K., Leggat, P.A., 2016. Human Monkeypox: Current State of Knowledge and Implications for the Future. Tropical Medicine and Infectious Disease 1, 8. https://doi.org/10.3390/tropicalmed1010008
- Osadebe, L., Hughes, C.M., Lushima, R.S., Kabamba, J., Nguete, B., et al., 2017. Enhancing case definitions for surveillance of human Monkeypox in the Democratic Republic of Congo. PLOS Neglected Tropical Diseases 11, e0005857. https://doi.org/10.1371/journal.pntd.0005857
- Reynolds, M.G., Damon, I.K., 2012. Outbreaks of human Monkeypox after cessation of smallpox vaccination. Trends in Microbiology 20, 80–87. https://doi.org/10.1016/j.tim.2011.12.001
- The Center for Food Security and Public Health, 2013. Monkeypox Technical Factsheet. http://www.cfsph.iastate.edu/Factsheets/pdfs/monkeypox.pdf (Accessed Mar 25, 2018).








