In the last two months, new SARS-CoV-2 strains have been identified in the United Kingdom, South Africa, and Brazil that harbor mutations associated with increased transmission within the population. Strain surveillance by rapid and constant NGS sequencing of positive samples allows for the identification of any mutants that pose a further threat to human health. Early knowledge of such threats is essential for an effective pandemic response and avoids overwhelming medical services around the world.
However, the global strain surveillance infrastructure is still in its infancy, meaning we are still largely in the dark about the epidemiological impact SARS-CoV-2 mutations are having. In this article, we zoom out on the last two months, highlighting where surveillance has shown success. We also underpin the increase in global strain surveillance efforts required in the coming months.
Surveillance successfully identifies new strains of concern
Toward the end of 2020, the UK government locked down the country to stem the increase in SARS-CoV-2 (COVID-19) infections. Scientists tracking the spread of the virus noticed that the southeast county of Kent was not following a typical trend in this lockdown period. While cases in the rest of the country were going down, cases in the southeast were rising.
On December 10th, Public Health England (PHE) first published the identification of a new Coronavirus strain, VoC-202012/01 (B.1.1.7), concentrated in Kent and parts of London. Retrospective studies showed cases originated in September. Current scientific understanding states that VoC-202012/01, which contains 23 mutations, has greatly increased transmissibility (it spreads faster). This strain can likely be attributed to the unusual increase in cases in the UK.
Epidemiological evidence shows that VoC-202012/01 doesn’t lead to a more severe SARS-CoV-2 infection. However, the increased transmissibility dramatically increases the risk of overwhelming health services and ultimately increases mortality rate. As of the 18th January, over 45 countries have reported at least one case of this variant.
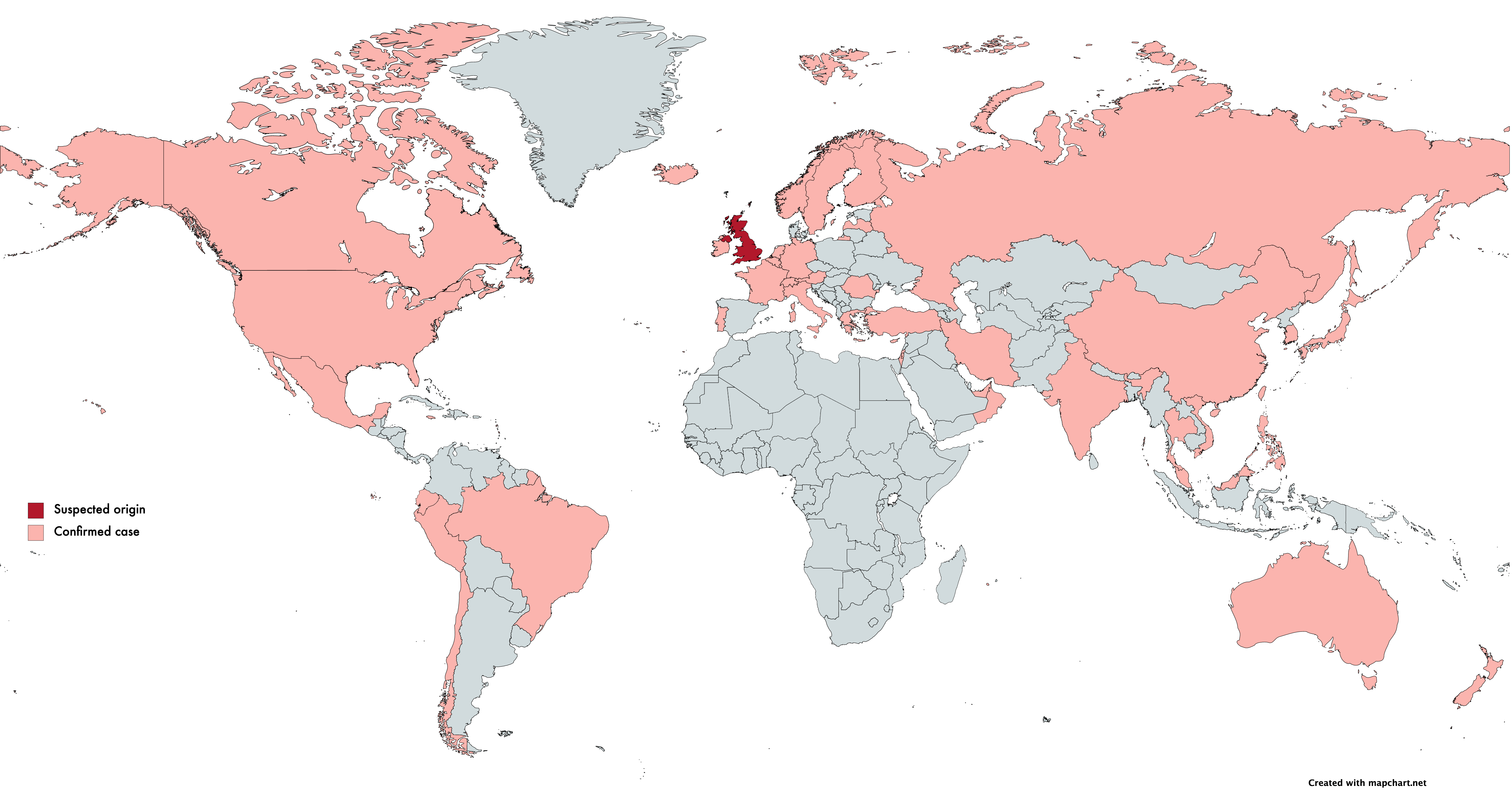
English scientists identified VoC-202012/01 by studying the routinely available SARS-CoV-2 sequencing data available due to the ongoing sequencing efforts in place in the UK through the COVID-19 Genomics UK consortium (COG-UK).
COG-UK formed in March 2020, consisting of hundreds of scientists across 16 sequencing hubs throughout the UK. To date, COG-UK has sequenced over 200,000 SARS-CoV-2 genomes, approximately 7% of the country’s positive tests. Before this, the largest dataset for epidemiological studies during an epidemic was 1,500 Ebola genomes. COG-UK surpassed this during their first month. COG-UK is currently aiming to sequence 20,000 genomes per week.
COG-UK has developed software that automatically analyzes the large volume of sequencing data produced, including the identification of mutations. Data is then distributed to the four public health institutions in the UK, which merge this with broader epidemiological data from PCR-based screening efforts. Mutations that have implications on public health are then identified, and advice is subsequently passed on to governing bodies. Positive strains are also submitted to the Global Initiative on Sharing Avian Influenza Data (GISAID), which has curated the global database of coronavirus genomes.
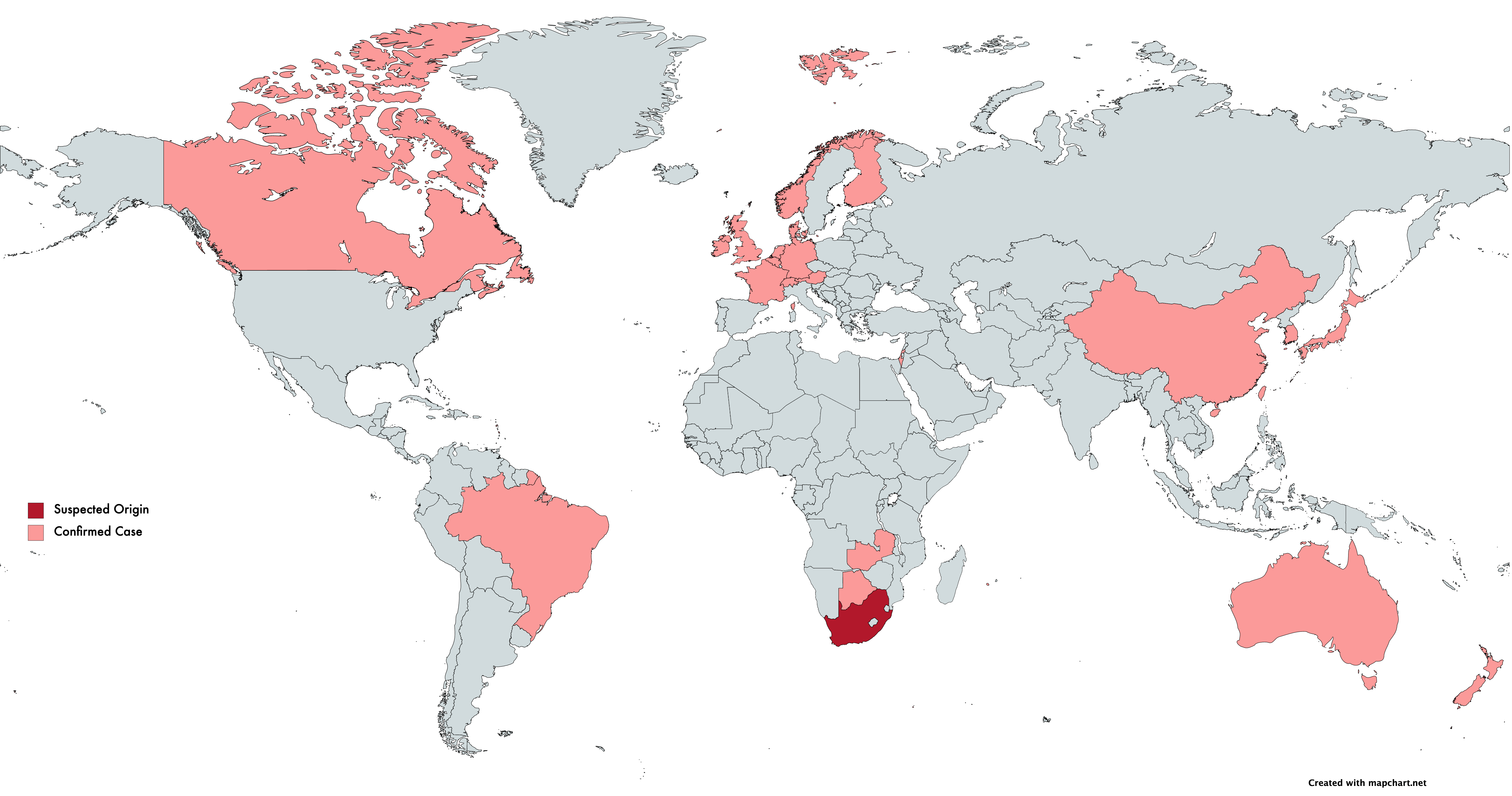
On December 21, 2020, the Africa Centers for Disease Control and Prevention announced that their surveillance work led to the identification of a unique coronavirus strain (501Y.V2). Three mutations were identified in the strain’s spike protein, one of which (N501Y) was also observed in the UK strain VoC-202012/01 and implicated in increased transmissibility. In light of 501Y.V2, The African CDC states that they are increasing surveillance efforts across Africa.
On January 6th, 2021, Japan’s National Institute of Infectious Diseases announced it had identified a new strain in four passengers traveling from Brazil, showing mutations implicated in increased transmissibility also present in VoC-202012/01 and 501Y.V2. A publication from the Instituto Leônidas e Maria Deane, Brazil, analyzed 148 genome sequences from positive cases in the Amazonas region. Results suggest this variant arose from the Amazonas region around December. With the recency of the strain, the authors state they are in the process of sequencing samples from new patients in the Amazonas region to understand the epidemiological impact.
We will only find variants if we are looking
During a pandemic, especially one of the scale and impact of SARS-CoV-2, the NGS sequencing of cases is essential to monitor changes in a viral genome for an effective pandemic response. Such surveillance is essential to “keeping ahead” of any new strains that pose an increased threat to human health. Understanding the scale and spread of these new strains allows governing bodies to allocate resources appropriately. As new strains are detected, one of the first and most salient questions to be asked is, “are current vaccines still effective?” For the UK and South Africa strains, early data suggests the answer is yes. Unfortunately, it is naive to assume this is the case for all strains in circulation when we have such a limited picture of sequence variation across the global population.
Logically, Japan, South Africa, and the UK have been among the first to identify coronavirus variants to date – all three nations have well-established sequencing infrastructure for strain surveillance. This begs two questions: what strains of concern may exist in countries that do not currently have adequate surveillance, and how far have these strains spread? A significant improvement to global strain surveillance infrastructure is required to provide answers.
Current WHO advice states that where feasible, countries with sequencing capacity should increase routine systematic sequencing of SARS-CoV-2 viruses from known cases to understand SARS-CoV-2 transmission patterns, and to monitor the emergence of new variants. Countries that do not have NGS capacity are encouraged to collaborate with public and private sequencing laboratories from the COVID-19 reference laboratory network. On January 8th, 2021, the WHO published comprehensive guidelines for the genomic sequencing of SARS-CoV-2 and the implementation of sequencing programs.
Recognizing the need for increased surveillance, the European Centre for Disease Prevention and Control has stated, “Member States need to perform timely genome sequencing of a significant and representative selection of isolates. Ideally, Member States should aim for similar timeliness, and fraction of samples sequenced [to the UK].” In a similar model to the UK response, positive cases will be reported to Europe’s Early Warning and Detection System and The European Surveillance System for epidemiological study.
The United States of America bears one of the most significant coronavirus surveillance challenges. In the first nine days of 2021, over two million cases were confirmed. Since the pandemic began, the US has submitted only 56,000 genome sequences of the 23 million US positive samples to GISAID. The country needs to screen three times this amount every week if it is to achieve surveillance numbers matching the UK and European goals. Currently, the CDC’s surveillance capacity is less than 3,000 genomes per week. The good news is efforts are underway to increase this capacity, and a surveillance network has recently been established.
Twist Bioscience’s contribution to the surveillance effort
Twist Bioscience recognizes the need for tools that enable the scientific community to perform rapid, efficient, and accurate sequencing of SARS-CoV-2 samples to identify variants. In November 2020, Twist launched its SARS-CoV-2 NGS Assay - RUO, a Research Use Only targeted sequencing assay designed to comprehensively hybridize with and capture the entire 30kb coronavirus genome from a patient sample.
In the WHO’s newly released advice on the sequencing of SARS-CoV-2 genomes, they say the following about target capture-based assays: “One advantage of using a capture-based approach... is that capture-based approaches can tolerate viral sequence differences from the probe sequences of 10–20%. This is higher than the mismatch tolerated by PCR, where such a divergence from the primer sequences would result in a high risk of amplicon failure. Capture-based approaches can therefore be used to enrich successfully for relatively divergent SARS-CoV-2 sequences.”
Twist offers a broad range of synthetic SARS-CoV-2 controls, allowing for verification of screens and sequencing assays and providing measures to determine the limit of detection and monitor test-to-test variance. Two controls for UK variant VoC-202012/01 are now available in response to the rapidly changing screening landscape.
Click the link for more information on tools for addressing the SARS-CoV-2 pandemic from Twist Bioscience.










