Twist Bioscience HQ
681 Gateway Blvd
South San Francisco, CA 94080
Sequence Design
How do I convert my Amino Acid sequence to a nucleic acid sequence?
Our website will accept amino acid sequences and convert them to a DNA sequence for synthesis. At the upload sequences page, select the Amino Acid Sequence option.
Make sure your input only contains Amino Acid one-letter characters. We support only the standard 20 one-letter code characters.
A "*" at the end of your amino acid sequences marks a stop codon (we do not add stop codons automatically).
Based on the Codon Usage Table you choose, the most frequently used codons will be selected.
If you do not see your specific organism, contact customersupport@twistbioscience.com for help.
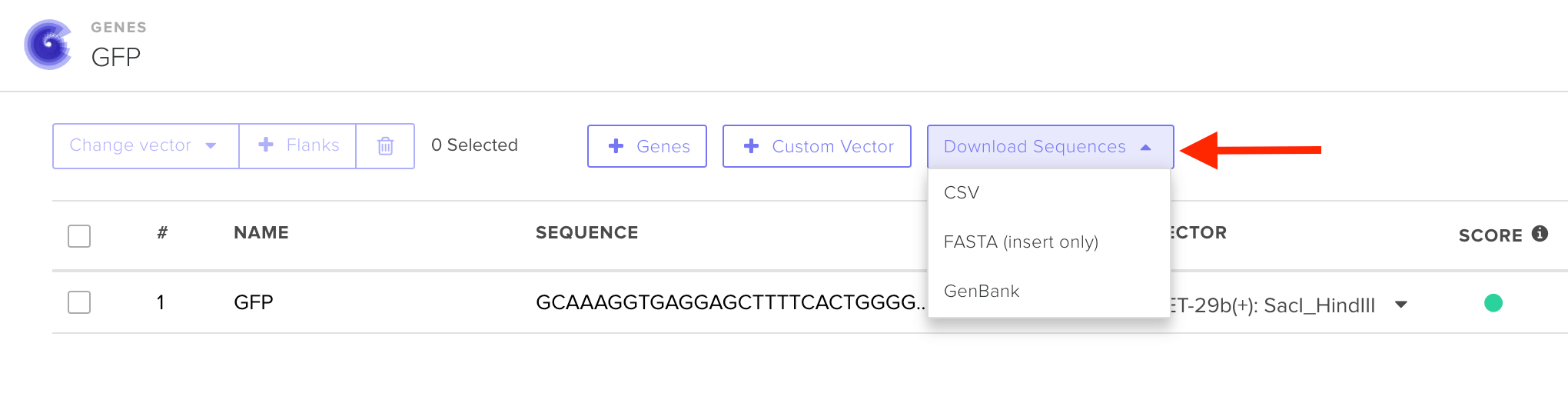
Note for Clonal Genes: Not all Twist expression vectors contain standard expression machinery upstream of the insertion point. We recommend double-checking the sequence of your final plasmid construct by clicking "Download Sequences" after selecting your desired vector.
Still have questions? Contact Us
What do the scoring results of my gene mean?
After submitting your gene sequences on our website, they will be automatically scored for feasibility of assembly:
Standard: No errors have been found. Gene sequence found to be of "standard" overall complexity (e.g. length, GC content, homology, etc).
Complex: No errors have been found. Some sequence complexities (e.g. length, GC content, homology, etc) exist but we do not expect any problems synthesizing your gene. Rarely, sequences may experience increased turnaround time or risk of manufacturing failure.
Error: Gene sequence contains errors. Please click on the sequence to get more details on how to resolve them.
Not Accepted: The sequence contains elements that make it impossible for us to synthesize. Please see the detailed explanations provided with the error message or refer to the “Design Guidelines”.
About Scoring System
Our scoring system adopts a machine learning model which analyzes and combines multiple sequence parameters ( i.e. overall GC percent, maximum homopolymer length, maximum repeat length, sequence length, repeat density, etc.). This model was trained using our historical manufacturing data, and it provides a more precise estimation of production success for genes and antibody products
About ISSUE messages
Warnings: Are associated with the risk of failure, predicted by the machine learning model. A higher number of warnings is more likely to result in a “Not Accepted” score.
Errors: Are not associated with gene complexity, and they can be fixed by correcting the gene design
Still have questions? Contact Us
What lengths of synthetic DNA does Twist currently offer for synthesis?
Customers have the option of ordering oligo pools, gene fragments, and clonal genes.
Oligonucleotides (linear single-stranded DNAs) in an oligo pool can range in size from 20 bp to 300 bp.
Gene fragments are linear double-stranded DNAs that can range in size between 300 bp and 5,000 bp in length.
Clonal genes are double-stranded DNAs that have been cloned into one of Twist’s vectors or a customer-provided custom vector. Clonal genes can range in size between 300 bp and 5,000 bp in length.
Still have questions? Contact Us
Are there any sequence limitations/design guidelines for genes which I should follow?
Synthesis issues are driven mainly by repetitive structures, extreme GC content, and homopolymers. We use a machine learning method to determine the chance of success. Therefore, most of the time, there is no single reason for the rejection of a sequence. Rather, complexity or rejection stem from a combination of features that push a sequence into a given bucket of complexity. The only hard rules are the following:
-
Avoid homopolymers >= 14bp
-
Do not include CcdB (Type II Toxin-antitoxin system)
Should you encounter an issue, we recommend removing or minimizing repeats, regions of extreme GC content, and homopolymers. We will highlight regions of concern if your sequence is complex, unbuildable, or contains sequences which conflict with our internal processes.
Still have questions? Contact Us
How do I convert my Amino Acid sequence to a nucleic acid sequence?
Our website will accept amino acid sequences and convert them to a DNA sequence for synthesis. At the upload sequences page, select the Amino Acid Sequence option.
Make sure your input only contains Amino Acid one-letter characters. We support only the standard 20 one-letter code characters.
A "*" at the end of your amino acid sequences marks a stop codon (we do not add stop codons automatically).
Based on the Codon Usage Table you choose, the most frequently used codons will be selected.
If you do not see your specific organism, contact customersupport@twistbioscience.com for help.
Note for Clonal Genes: Not all Twist expression vectors contain standard expression machinery upstream of the insertion point. We recommend double-checking the sequence of your final plasmid construct by clicking "Download Sequences" after selecting your desired vector.

Still have questions? Contact Us
What do the scoring results of my gene mean?
After submitting your gene sequences on our website, they will be automatically scored for feasibility of assembly:
Standard: No errors have been found. Gene sequence found to be of "standard" overall complexity (e.g. length, GC content, homology, etc).
Complex: No errors have been found. Some sequence complexities (e.g. length, GC content, homology, etc) exist but we do not expect any problems synthesizing your gene. Rarely, sequences may experience increased turnaround time or risk of manufacturing failure.
Error: Gene sequence contains errors. Please click on the sequence to get more details on how to resolve them.
Not Accepted: The sequence contains elements that make it impossible for us to synthesize. Please see the detailed explanations provided with the error message or refer to the “Design Guidelines”.
About Scoring System
Our scoring system adopts a machine learning model which analyzes and combines multiple sequence parameters ( i.e. overall GC percent, maximum homopolymer length, maximum repeat length, sequence length, repeat density, etc.). This model was trained using our historical manufacturing data, and it provides a more precise estimation of production success for genes and antibody products
About ISSUE messages
Warnings: Are associated with the risk of failure, predicted by the machine learning model. A higher number of warnings is more likely to result in a “Not Accepted” score.
Errors: Are not associated with gene complexity, and they can be fixed by correcting the gene design
Still have questions? Contact Us
What lengths of synthetic DNA does Twist currently offer for synthesis?
Customers have the option of ordering oligo pools, gene fragments, and clonal genes.
Oligonucleotides (linear single-stranded DNAs) in an oligo pool can range in size from 20 bp to 300 bp.
Gene fragments are linear double-stranded DNAs that can range in size between 300 bp and 5,000 bp in length.
Clonal genes are double-stranded DNAs that have been cloned into one of Twist’s vectors or a customer-provided custom vector. Clonal genes can range in size between 300 bp and 5,000 bp in length.
Still have questions? Contact Us
Are there any sequence limitations/design guidelines for genes which I should follow?
Synthesis issues are driven mainly by repetitive structures, extreme GC content, and homopolymers. We use a machine learning method to determine the chance of success. Therefore, most of the time, there is no single reason for the rejection of a sequence. Rather, complexity or rejection stem from a combination of features that push a sequence into a given bucket of complexity. The only hard rules are the following:
-
Avoid homopolymers >= 14bp
-
Do not include CcdB (Type II Toxin-antitoxin system)
Should you encounter an issue, we recommend removing or minimizing repeats, regions of extreme GC content, and homopolymers. We will highlight regions of concern if your sequence is complex, unbuildable, or contains sequences which conflict with our internal processes.
Still have questions? Contact Us








