PEGS 2018: The Backdrop for Advancement in Variant Library Technology

PEGS (Protein Engineering Global Summit) is the largest annual event of its kind, covering all aspects of the current state of biological drug development, including protein and antibody engineering. This year’s conference in Boston, MA lived up to its billing, with more than 2,500 industry professionals in attendance.
Taking place in early May, the conference hosted over 400 lectures, panels, tutorials and round-table discussions, along with more than 150 exhibitors, and 300 research posters on display in the exhibit hall. The array of distinguished faculty boasted some of the industry’s leading experts, visionaries, and influencers from top pharma, biotech, academia, and government institutions.
Twist Bioscience was there to continue spreading the word about the value of its burgeoning Variant Libraries technology through a User Group Meeting, exhibit booth and scientific poster presentation. Twist Bioscience is building a community of users committed to using their next generation DNA synthesis-powered Variant Libraries in order to push forward the protein engineering industry through the use of advanced technologies.
This community gathered at PEGS in Twist’s Library User Group Meeting, hosted by Garry Merry, Twist’s Director of Library Business. “What we’d like to do tonight is let our users and collaborators really introduce their technology that uses front-end Twist Combinatorial Libraries to you,” Merry said, kicking off the event.

Garry Merry from Twist Bioscience was the master of ceremonies at the Twist Bioscience Libraries User Group Meeting at PEGS Boston.
Lucy Jia Xu, Twist Bioscience’s Research Associate, spoke first at the event. She outlined how Twist Bioscience is providing industry leading access to sequence variation with the next generation of synthetic gene libraries available to the protein engineering market. She explained, “[E]very library is built from a starting substrate of ultra-high quality oligonucleotides. The variation in the library is directly fabricated individually, codon-by-codon.” This means that everything a customer designs is precisely what is delivered. Every variant library fabricated by Twist is QC’d with NGS. “We are able to offer precision, user defined composition, and next generation sequencing validation so that you can focus your libraries and push the boundaries of what you can explore in the variant space,” Lucy explained.

Michael R. Dyson, PhD, CTO of IONTAS
Michael R. Dyson, PhD, the CTO of IONTAS spoke next. He explained how IONTAS is using Twist Bioscience’s Libraries to enable mammalian display-based antibody discovery, a technology Merry dubbed the “holy grail of antibody engineering.”
“So what’s the advantage of mammalian display?” Dyson asked the audience. “It allows us to produce the antibody screen with the same post translation and modifications that you would have in a final production therapeutic.”
“So we have a case study where we had a starting anti-PD1 antibody that was functionally active, but had poor affinity. We used the combination of mammalian display with a Twist library to take this antibody through the affinity maturation process,” Dyson added. “With Twist we could produce a CDR3 library on the heavy and light chains, giving us 108 possible variants, a size we can easily display on mammalian cells. If we wanted to produce that with traditional library generation methods we would need a library of 1024 variants, which is impossible.”
“From this library we were able to get highly functional antibodies straight off from the mammalian display selection. Seventeen out of 23 of our top hits were more potent than Nivolumab, the clinically approved anti-PD1 antibody.”

Jacob Glanville, PhD, CSO of Distributed Bio
Distributed Bio’s Co-Founder and CSO, Jacob Glanville, PhD, followed. He showed how his company rationally engineered antibody discovery technologies for the highly challenging target family of G-protein coupled receptors, using Twist Bioscience’s Libraries.
“I try to use deep sequencing and deep functional data recovery to try to inform and optimize iterative design improvements on antibody discovery,” Glanville said in his presentation. “Throughout the entire history of my work, I’ve always been limited. My imagination of what I could produce has been limited by the synthesis technologies.”
“That’s why I was so ecstatic when I started seeing what Twist was capable of,” Glanville added. “It really was the most exciting thing that’s happened in the space of synthetic immunology, and really synthetic biology engineering. Because it enabled certain types of designs that previously were not possible.”

Ryan Kelly, PhD, Research Scientist at xCella Biosciences
Ryan Kelly, PhD, a research scientist at xCella Biosciences, closed the show. He outlined how the xCella platform enables high-throughput functional screening of Twist Bioscience antibody libraries.
“We came to Twist with a problem and we wanted to make a very high quality antibody library that recapitulated as best as possible, what you see in the human repertoire,” Kelly told the audience. “Except we had some design considerations. We asked very specifically for framework pairings that would be amenable to highly developable antibodies, and we designed an H3 segment which had quite a few specifications in it, including eliminating motifs for non-specificity and developability, and eliminating other common liabilities which you might see in many first generation synthesis libraries.”
“So Twist was able to build this for us in a full scFv (single-chain variable fragment) format, which really allowed us to accelerate our discovery process and build this library very quickly once it was synthesized for us,” Kelly added.
From the various talks at the meeting, it’s clear that access to high quality variant libraries is essential for the current state of protein and antibody engineering. When we spoke to Merry afterward about his reflections on the meeting, he said: “It really showed us that people are beginning to realize that degenerate oligo-based methods are not optimal for the current state of the protein engineering field.”
Using traditional library fabrication methods, including those that use degenerate oligos, bias is introduced into the sequence space. “If you do the math, you can see that this bias causes a considerable disconnect between theoretical diversity and delivered diversity in the variant library,” Merry explained. “Professor Manfred Reetz recently published excellent empirical data on the issues that bias in the library can cause for protein engineering experiments.” You can read the paper Merry described here.
Merry continued, stating, “This bias meant users had to screen traditional libraries multiple times to confirm whether it contained useful variants. Screening used to be 10x the cost of the library.” Now, with the new library technology provided by Twist Bioscience, this bias is removed, and the screening effort required is significantly reduced. Twist Bioscience presented a scientific poster at PEGS, that can be viewed here, showing data that details how bias is eliminated from their libraries.
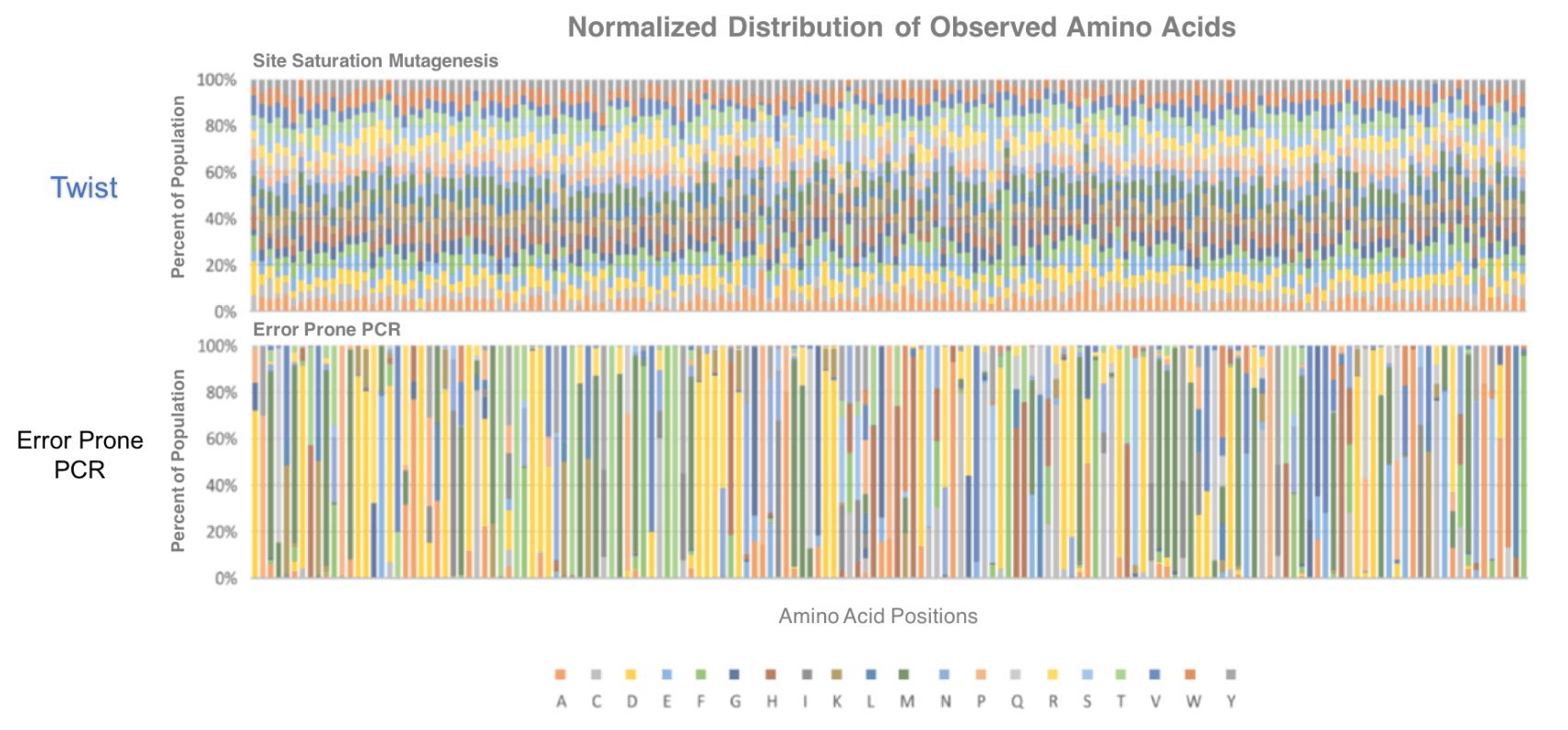
The ratio of variants observed in a directly synthesized site saturation mutagenesis library provided by Twist, verses the variants observed with an equivalent library generated by error-prone, PCR-based methods.
Simply, this bias is removed because every variant is synthesized directly by Twist Bioscience. As variation is directly synthesized, users have complete flexibility of design. For example, customers are able to generate a library of antibodies containing variation in both the heavy and light chain at all three CDRs, with up to 1010 variants. Variation can be introduced as single or multiple saturated sites, as binary substitutions, as specific ratios of amino acids, as regions of varied sequence length, or as combinations of multiple pre-existing germlines.
When asked to share his thoughts on what the future holds for protein engineering now that customers can access such high quality tech, Merry shared that, “Part of Twist’s future involves continuing to work with our growing community to expand discovery and development in the protein engineering and antibody field.”
Merry continued, saying, “Right now, many people in the antibody engineering field still rely on natural immune systems that are challenged with a given antigen to generate their antibody sequence variation. This is a laborious and imprecise process. In the future, now that Twist Bioscience has made high quality libraries so accessible, more and more research will move away from the use of natural diversity.” Such a move is set to dramatically accelerate and simplify the antibody discovery process.
Twist Bioscience is looking forward to continuing to support our growing Library User community at the forefront of advancement in protein and antibody engineering. We will meet with the community again at the next installment of the PEGS conference in Lisbon, Portugal in November.
What did you think?
Like
Dislike
Love
Surprised
Interesting