Twist Bioscience recently partnered with New England Biolabs (NEB®) to create an end-to-end methylation sequencing (methyl-seq) workflow for the improved detection of genomic methylation patterns. In this article, we dive into how this pipeline leads to increased confidence in methylated-base calls for the study of epigenetics.
Among the most studied epigenetic modifications, DNA methylation occurs when DNA methyltransferases add a methyl group to the cytosine residue of cytosine-phosphoguanine (CpG) dinucleotides. This modification contributes to a wide variety of biological processes by promoting genomic instability or transcriptional silencing. Affected processes include normal processes, like cellular differentiation, as well as abnormal ones, like carcinogenesis.
Oncologists have considered a number of candidates in their search for diagnostic biomarkers, but few are as compelling as DNA methylation. For one, aberrant DNA methylation occurs very early during cancer progression. DNA methylation also manifests inconsistent patterns across the genome, making it possible to assay multiple loci for a more specific and sensitive test. For these reasons, detecting DNA methylation is an attractive approach for early cancer screening, particularly when it can be done non-invasively, as in liquid biopsies of circulating tumor DNA (ctDNA).
Detecting methylation by sequencing is not without its share of challenges. The methyl-seq procedure can be difficult to apply to the whole genome without starting with a substantial amount of sample DNA. Methyl-seq also complicates efforts to reduce cost by target enrichment because libraries converted for methylation detection are inherently more difficult to capture. Below, we discuss how the new Twist methyl-seq workflow clears these hurdles to provide a cost-effective, yet highly efficient, alternative to existing methyl-seq workflows.
Historically, bisulfite sequencing has been considered the gold standard for DNA methylation detection. This technology uses bisulfite salts to chemically convert unmethylated cytosines into uracils. Subsequent PCR amplification replaces these uracils with thymines. When the resulting libraries are sequenced by next-generation sequencing (NGS), originally unmethylated and methylated cytosines are sequenced as thymines and cytosines, respectively.
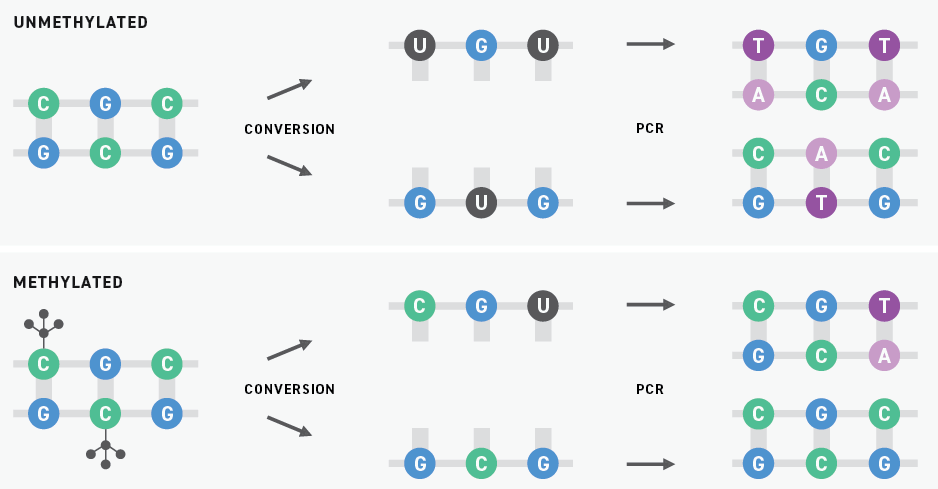
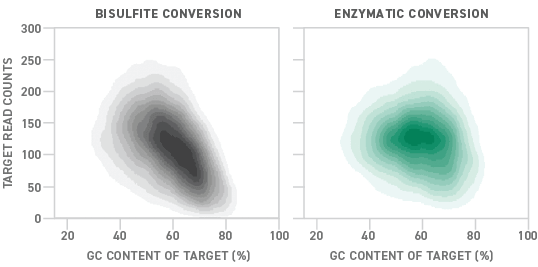
Although bisulfite sequencing provides a quantitative readout with high throughput and single-base pair resolution, bisulfite treatment can degrade DNA samples substantially. Also, because unmethylated cytosines are particularly prone to degradation, bisulfite conversion can lead to a drop-off in coverage of regions with high GC content. Sample degradation reduces the sensitivity of the assay, making it more difficult to apply it to samples that are inherently low in concentration, like ctDNA.
NEB recently pioneered an enzymatic alternative to the conventional bisulfite conversion process. Called enzymatic methyl-seq (EM-seq™), the new methyl-seq library preparation method uses a series of enzymes to convert DNA samples for methylation detection. Compared to bisulfite-converted libraries, those prepared enzymatically show more uniform coverage across a variety of GC contents. This translates into a 15% higher detection rate for EM-seq than bisulfite conversion.
As with many NGS assays, the overall cost and sequencing effort can be reduced by target enrichment. This step can be inserted before or after conversion in methylation sequencing workflows depending on the amount of input sample DNA available. For certain research applications where sample DNA is easy to come by, target genomes can be enriched before conversion. This route poses fewer problems to panel design, as it only needs to account for unconverted DNA sequences.
Target enrichment can be performed after conversion to enable more sensitive applications, but there’s a catch (or two): Conversion generates additional, unique DNA sequences that also need to be factored into the panel design. Conversion also simplifies DNA sequences by converting many cytosines to thymines. Such sequences can be more difficult to capture, which is why many targeted methyl-seq experiments suffer from high off-target rates.
To overcome the challenges of pre-capture conversion, Twist leaned on its expertise in custom panel design and its massively-parallel synthesis platform. The result: Twist Custom Methylation Panels. Optimized for methylation detection, these panels include probes to capture all 4 potential sequences at a given site: methylated, unmethylated, sense and antisense. Different stringencies can also be selected to further optimize capture performance in a given experiment. Being built on the Twist DNA Synthesis Platform, Twist custom Methylation Panels offer the same fidelity, uniformity, and flexibility as Twist Custom Panels.
Additionally, Twist has developed a broad, off-the-shelf methylation capture panel known as the Twist Human Methylome Panel. This carefully designed panel provides coverage over more than 3.2 million CpG sites, enabling enrichment for 84% of CpG islands in the human genome as well as several million additional biologically relevant methylation loci. When paired with Twist’s NGS Methylation Detection System, this panel can deliver highly uniform, precise, and broad methylation sequencing data.
The combination of EM-seq and Twist Custom Methylation Panels (custom or off-the-shelf) provide an end-to-end solution for targeted methyl-seq that performs well across the gamut of methylation levels. Rounding out the all-in-one solution is Methylation Enhancer, a methyl-seq-specific hybridization blocker that minimizes off-target capture when combined with a Twist Custom Methylation Panel. The entire solution is called the Twist NGS Methylation Detection System.
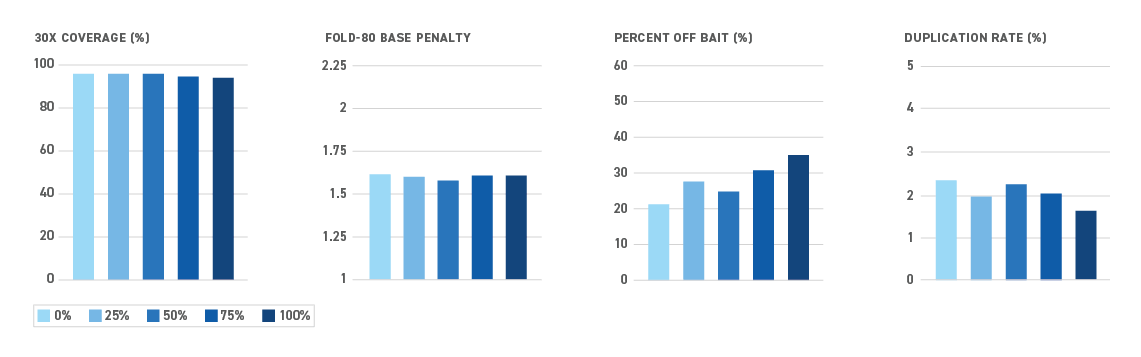
The Twist NGS Methylation Detection System offers the most efficient targeted methyl-seq solution yet — and all in one box. By reducing sample loss during library prep and hybrid capture, the system enables even the most sensitive applications.
Learn more about Twist Methylation Detection System here, or contact us.
您怎么认为?
喜欢
不喜欢
喜爱
惊讶
有兴趣
