エラーの少ない NGS ライブラリ調製でシーケンシング品質を改善

In this article, we highlight how quality library preparation reduces sequencing error rates while providing high-yield library preparation. Twist has just upgraded its suite of library preparation tools by creating the Library Preparation Enzymatic Fragmentation (EF) Kit 2.0. We detail how this kit has been built to help you achieve better quality library preparation for better sequencing results.
Errors are inevitable when preparing DNA libraries for Next-Generation Sequencing (NGS), particularly when starting with low volume samples. Technical limitations mean that base misincorporations and inappropriate recombinations or ligations are likely to occur. Such errors can be costly for any experiment, particularly when attempting to discover rare variants or when working with limited sample input volumes.
In these situations, where efficiency and accuracy are critical to meaningful NGS results, the library preparation kit you choose matters.
Here, we’ll dive into the Twist Library Preparation EF Kit 2.0—an upgraded library preparation kit that enables efficient library preparation from fragmentation to amplification, and reduces error rates to help you generate more accurate NGS results.
Fewer Chimeras During Library Preparation
When preparing a sequencing library for NGS, it’s necessary to ligate sequencing adaptors to the DNA fragments. However, the process of ligating adaptors has inherent limitations that sometimes result in ligation of disparate DNA fragments to one another, forming what’s known as a chimera2. These structures can also form during amplification of the library when sequences are similar enough to prime off of one another.
Chimera’s are simply defined as sequencing reads that map to two different, non-overlapping locations in the target DNA. A chimera can be a legitimate sequence that exists within your sample, usually as a product of structural variations (such as DNA translocations). Or, chimeras can be sequencing library artifacts, formed during library preparation when DNA fragments inappropriately recombine during amplification or concatenate during ligation, resulting in an artificial sequence2.
The latter is a significant hurdle in the clinical setting when diagnosing patients who may well harbor structural variations, as in the case of Ewing Sarcoma3. In these situations, proving the legitimate presence of a chimera can be difficult. By lowering the percentage of artificial chimeras formed during library preparation, you can greatly decrease your false-positive rate and minimize wasted reads.
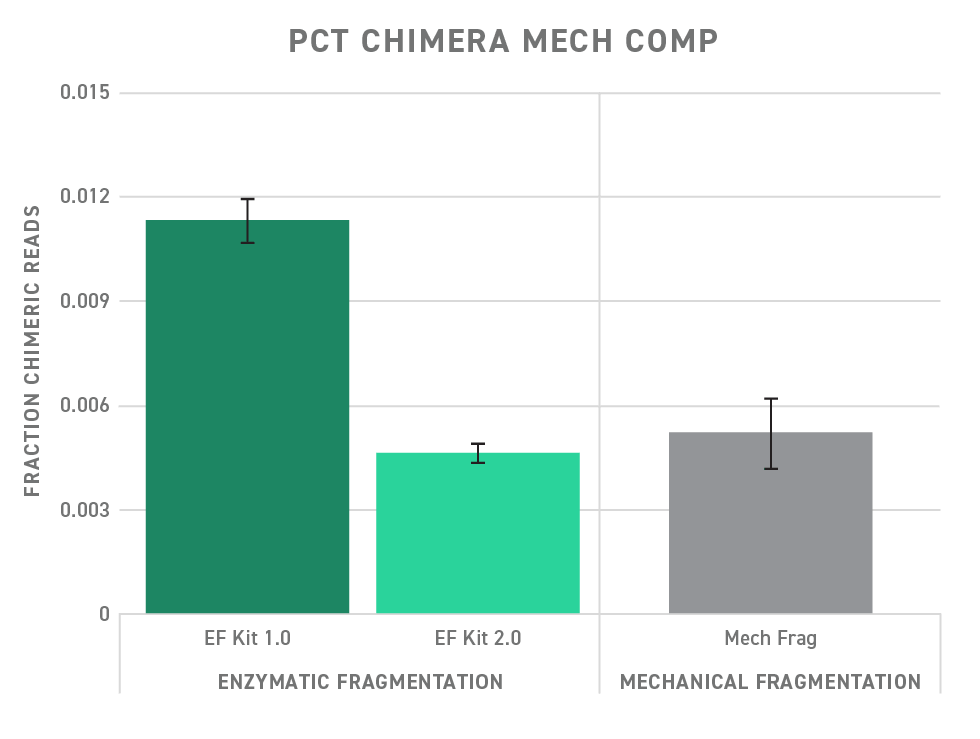
The upgraded Twist Library Preparation EF Kit 2.0 helps you minimize chimeras by providing an optimized reaction mixture for faithful ligation. As a result, the Twist Library Preparation EF Kit 2.0 shows a significantly decreased fraction of chimera reads compared to early versions of the Twist Library Preparation EF Kit (version 1.0). Performance, with respect to chimera formation, is improved to the point of being on par with mechanical fragmentation.
Decrease Sequencing Errors During Library Amplification
Base misincorporation is an inherent problem during library amplification because it creates artificial mutations that may later be interpreted as bona fide variants. This is particularly problematic when studying rare variants, or when working with heavily damaged DNA, such as that found in archaic specimens or formalin-fixed paraffin-embedded (FFPE) samples4.
Improving amplification accuracy requires optimization of amplification conditions and selection of the right polymerase.
To that end, the Twist Library Preparation EF Kit 2.0 contains the Equinox Polymerase Mix. This mix contains a hot-start, high-fidelity polymerase with a broad tolerance for GC content (ranging from 20 - 80%), enabling robust, high-yield, and accurate library preparation.
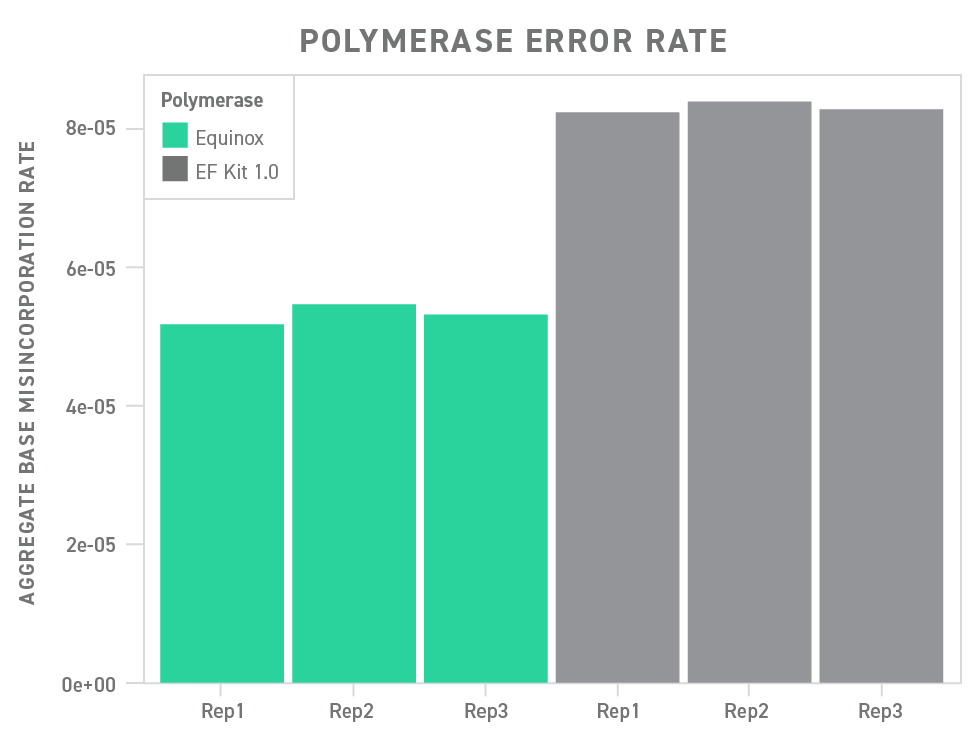
Underscoring this point, the polymerase included in the Twist Library Preparation EF Kit 2.0 outperforms a commonly used polymerase in a head-to-head comparison, showing a significant reduction in base misincorporation.
Efficient Target Enrichment
Enriching your library for target sequences allows you to focus your resources and avoid over sequencing non-target regions. Efficient target enrichment for NGS library preparation requires uniform target capture.
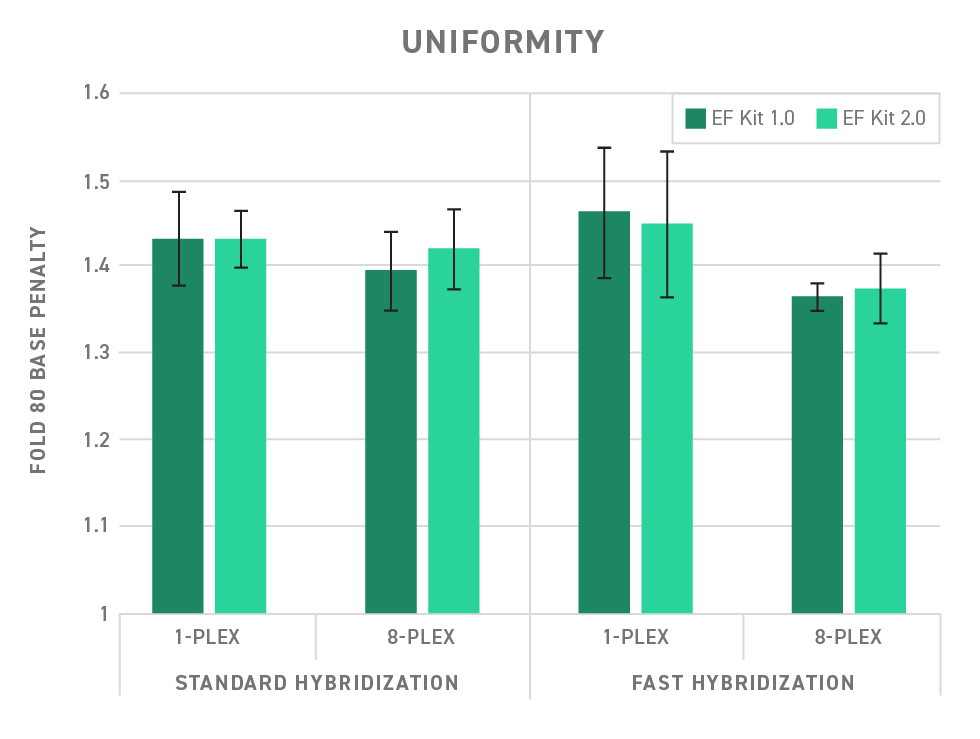
Uniformity refers to how even target capture occurs. Without uniformity, target enrichment is more likely to be biased toward the capture of one sequence over another. In effect, this means many more rounds of sequencing would be needed to ensure that poorly captured targets are adequately represented. This is summarized as a fold-80 score, which describes how much over-sequencing must be performed to bring 80% of the sequences up to mean coverage.
When the fold-80 score is high, you begin to accumulate costly, wasted sequencing reads for the well-captured targets. Therefore, lowering the fold-80 score can be cost-effective as you reduce the amount of sequencing needed to assess each target sequence adequately. Read our white paper for an in-depth review of the importance of uniformity in target capture.
The Twist Library Preparation EF Kit 2.0 easily integrates with Twist target enrichment workflows, enabling a high level of uniformity. (See Figure 3)
Low sample volume? Go with low risk NGS library prep
Putting it all together, the Twist Library Preparation EF Kit 2.0 empowers you to make the most out of your samples, even when you’re working with challenging samples like liquid biopsies. The kit’s streamlined workflow combines library preparation steps into a single-tube reaction which, alongside a low chimera rate and Twist’s highly uniform target enrichment workflow, translates into better quality library preparation and ultimately sequencing results you can trust.
Click here to learn more about the Twist Library Preparation EF Kit 2.0 and other library prep kits.
参考文献
- Schwarzenbach, Heidi, et al. “Cell-Free Nucleic Acids as Biomarkers in Cancer Patients.”Nature Reviews Cancer, vol. 11, no. 6, 2011, pp. 426–437., doi:10.1038/nrc3066.
- Haas, Brian J et al. “Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons.”Genome research vol. 21,3 (2011): 494-504. doi:10.1101/gr.112730.110
- Delattre, Olivier, et al. “The Ewing Family of Tumors -- A Subgroup of Small-Round-Cell Tumors Defined by Specific Chimeric Transcripts.”New England Journal of Medicine, vol. 331, no. 5, 1994, pp. 294–299., doi:10.1056/nejm199408043310503.
- Stiller, M., et al. “Patterns of Nucleotide Misincorporations during Enzymatic Amplification and Direct Large-Scale Sequencing of Ancient DNA.”Proceedings of the National Academy of Sciences, vol. 103, no. 37, 2006, pp. 13578–13584., doi:10.1073/pnas.0605327103.
いかがでしたか?
気に入った
気に入らなかった
大好き
驚かされた
面白かった
