Capturing The Basics of NGS Target Enrichment

With limited time and resources, researchers often have difficult decisions to make, particularly when it comes to sequencing. Though the costs and efficiency of next generation sequencing (NGS) have improved considerably in recent years, the process remains a substantial drain on laboratory resources. As such, researchers have to be judicious in their studies, balancing the depth, breadth, and scale of sequencing in order to get the most discovery bang for their buck.
One of the most common ways to find this balance is through target enrichment—a method wherein specific segments of DNA or RNA are isolated prior to sequencing, enabling the exclusion of unwanted genetic loci from the sequencing process. With fewer loci to sequence, researchers can reallocate resources to achieve deeper, higher-quality data. In the sections that follow, we’ll explore the concept of target enrichment and detail critical considerations when designing your experiment.
The Tension Between Depth, Breadth, and Scale
When designing an NGS experiment, every researcher must consider the depth, breadth, and scale of sequencing that’s needed in order to answer their specific questions.
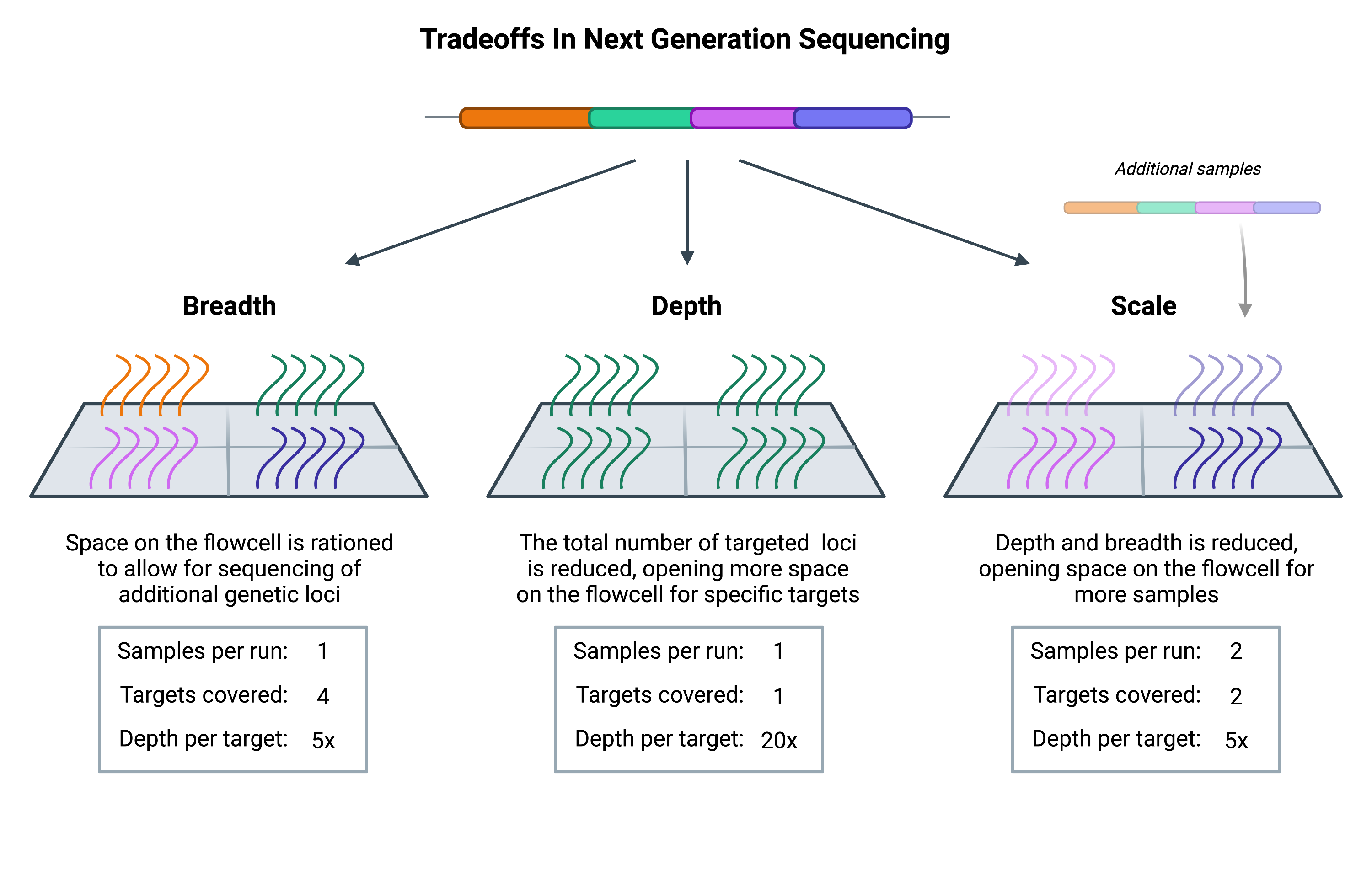
Breadth describes how much of the genome (or transcriptome) is sequenced, while depth is a measure of repetition, describing the number of times that a specific genetic locus is sequenced. Scale refers to how many samples can be run. With greater depth comes greater confidence that the data is accurate and not obscured by sequencing errors. However, achieving greater depth means that each target sequence will occupy more space on the NGS flowcell (the physical structure where sequencing takes place).
With only a finite amount of space per flowcell, increasing the depth of sequencing will require free space on the flowcell. Such space can be made by either decreasing the number of genetic loci, or by decreasing the number of samples that are loaded (thus decreasing the breadth and scale, respectively).
Whether you choose to invest in depth, breadth, or scale depends on the questions you’re asking. Researchers carrying out population-level studies often have a large number of samples to process and may need to increase scale by reducing the breadth or depth of sequencing to compensate. On the other hand, those studying mendelian disorders, which are primarily associated with specific mutations in single genes, may have little concern over breadth and are instead focused on depth vs scale.
The costs associated with whole genome sequencing are such that most projects cannot afford to reach the depth or scale needed to carry out their study. Therefore they have to reduce the breadth, and they do so through target enrichment.
Target Enrichment: Hybridization vs Amplicons
The basic premise behind target enrichment is that DNA libraries can be modified so that specific genetic loci are deliberately overrepresented in the sample pool prior to sequencing. Historically, this was done using amplicon-based sequencing wherein PCR amplification is used to selectively and extensively amplify target sequences. One of the benefits of this approach is that it allows low quantities of DNA to be preferentially amplified, creating many repeat molecules for each targeted locus. This makes it possible to achieve deeper coverage despite having very few DNA molecules in the original sample.
However, with this benefit comes the risk of artificial variation. Even the best polymerases can make mistakes that lead to variant DNA sequences. If these mistakes happen early in the amplification process, researchers may have difficulty recognizing the error, ultimately degrading confidence in the resulting data. Additionally, it’s difficult to multiplex (run multiple tests on a sample at once) and scale amplicon-based target capture—primer-dimers and the nuanced challenges of PCR probe design compound as the number of targets increases.
Though high-fidelity polymerases have been developed to mitigate this risk of amplification errors, the limitations of amplicon target enrichment have diminished its application in clinical and research settings. Instead, many have turned to hybridization-based approaches (also referred to as hybrid capture).
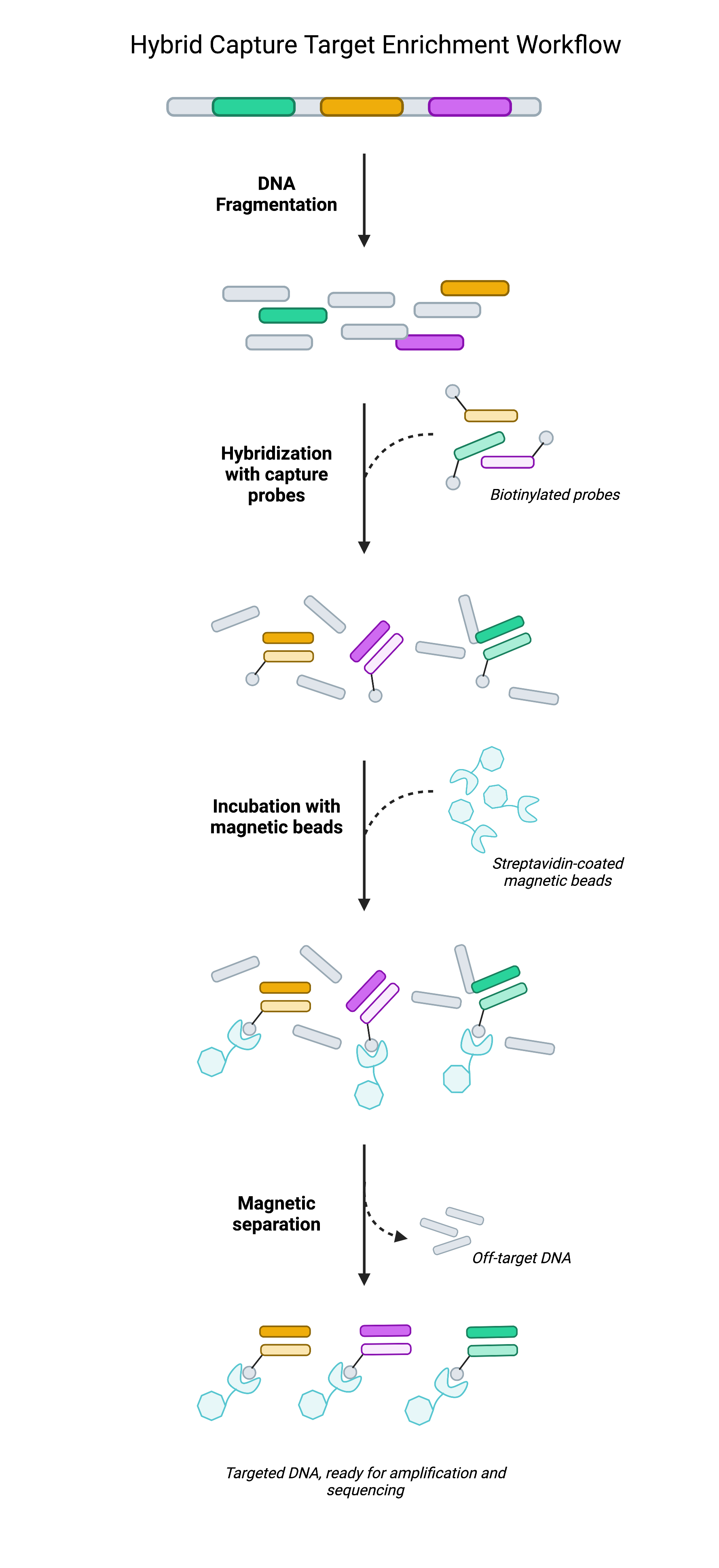
Rather than rely on selective PCR amplification, hybrid capture techniques achieve target enrichment by physically isolating target regions prior to sequencing. Hybrid capture often leverages biotinylated oligonucleotide probes (short pieces of DNA) that are designed to be complementary to genomic regions of interest. Once bound to target regions, the target-probe complex can be immobilized on streptavidin-coated beads as the remainder of the DNA library—the regions not being targeted—are washed away. What remains is a focused DNA library largely composed of target regions.
In isolating select portions of DNA, researchers have effectively reduced the breadth of DNA that’s loaded onto the flowcell from each sample. With less space occupied by each sample, there is more room to work within each sequencing run. This extra space can then be used to improve sequencing depth (by loading higher volumes of each sample) or else to increase the scale of the study (by loading more samples per flowcell). As the cost of sequencing is largely dictated by the number of sequencing runs performed, having the ability to maximize the number of samples on a flowcell helps researchers reduce costs without sacrificing data quality1.
Head-to-head comparisons of amplicon and hybrid capture approaches highlight the benefits and limitations of both. Hybrid capture tends to require more technical expertise and may take longer than amplicon sequencing to perform. Still, hybrid capture achieves more comprehensive target capture, better uniformity of coverage, and greater analytical sensitivity2.
Real-world applications of target capture
Put simply, hybrid capture is an effective way to focus resources where they’re needed most. But the simplicity of this statement obscures the significant impact that target capture can have in both clinical and research settings.
For example, single-cell RNA sequencing is increasingly used to explore the diversity of cell states within tissues. Such studies can provide valuable insight into tumor evolution, tissue development, stem cell differentiation, and more. However, these studies are limited in their ability to detect lowly expressed genes, including many transcription factors, whose minor signal can be mistaken for noise (this phenomenon is known as a “dropout”). As a result, significant biological data is likely to be overlooked, unless well-designed target capture panels are used.
The benefits of custom target capture probes for single-cell RNA sequencing was emphasized in a recent study by Replogle et al, wherein the authors aimed to sequence 978 of the landmark L1000 genes. While target transcripts initially represented only 6% of sequenced molecules, target capture enriched the sequencing data for targets 14-fold, such that 87% of sequenced transcripts were those being targeted by the researchers3.
Similarly, target capture can be critical when attempting to detect circulating tumor DNA (ctDNA). These tiny fragments of tumor-derived DNA can be powerful biomarkers in minimal residual disease testing (MRD), but detecting ctDNA is exceedingly difficult owing to their rarity among other cell-free DNA (cfDNA) molecules. Following curative treatment, ctDNA may represent as little as 0.01% of the total cfDNA present in a liquid biopsy sample. It is easy for ctDNA to be lost as dropouts in this scenario. However, target capture probes designed to match tumor-specific sequences can substantially increase the sensitivity of these assays, enabling earlier detection of tumor recurrence4,5.
In short, target capture can be an invaluable tool for a wide range of sequencing projects. In the following chapter, we’ll explore some key considerations when selecting or designing a target capture panel.
参考文献
- Samorodnitsky, Eric et al. “Evaluation of Hybridization Capture Versus Amplicon-Based Methods for Whole-Exome Sequencing.”Human mutation vol. 36,9 (2015): 903-14. doi:10.1002/humu.22825
- Singh, Rajesh R. “Target Enrichment Approaches for Next-Generation Sequencing Applications in Oncology.”Diagnostics (Basel, Switzerland) vol. 12,7 1539. 24 Jun. 2022, doi:10.3390/diagnostics12071539
- Replogle, Joseph M et al. “Combinatorial single-cell CRISPR screens by direct guide RNA capture and targeted sequencing.”Nature biotechnology vol. 38,8 (2020): 954-961. doi:10.1038/s41587-020-0470-y
- Larribère, Lionel, and Uwe M Martens. “Advantages and Challenges of Using ctDNA NGS to Assess the Presence of Minimal Residual Disease (MRD) in Solid Tumors.”Cancers vol. 13,22 5698. 14 Nov. 2021, doi:10.3390/cancers13225698
- Kurtz, David M et al. “Enhanced detection of minimal residual disease by targeted sequencing of phased variants in circulating tumor DNA.”Nature biotechnology vol. 39,12 (2021): 1537-1547. doi:10.1038/s41587-021-00981-w
いかがでしたか?
気に入った
気に入らなかった
大好き
驚かされた
面白かった