Banques à haute diversité, construites
de manière experte pour votre cible
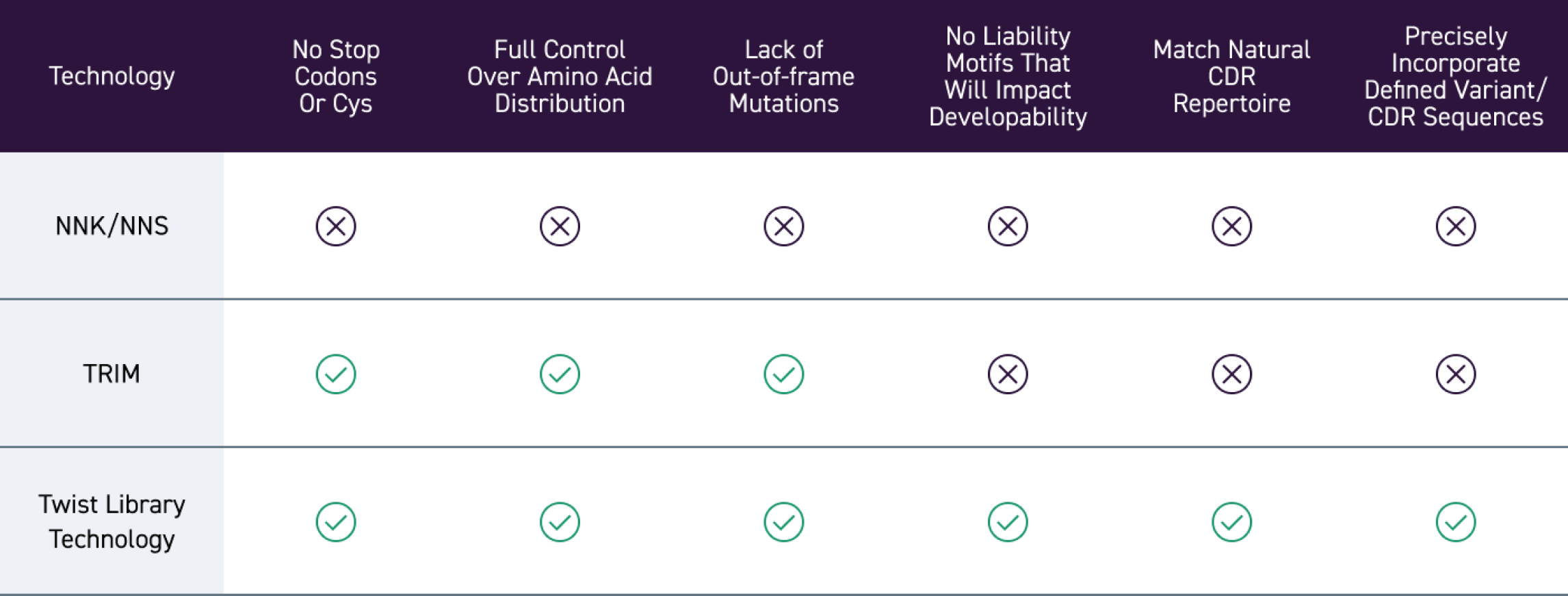
Les méthodes de construction de banques sur mesure traditionnelles peuvent produire des codons redondants, une diversité imprévisible ainsi qu’une distribution d’acides aminés non uniforme.
Avec une précision base par base, notre plateforme de synthèse d’ADN sur silicone permet un contrôle précis de la diversité des banques, du rapport de distribution des acides aminés, de la variation de longueur des CDR, ainsi que de l’utilisation des codons, ce qui permet la construction de banques sur mesure de haute qualité, se concentrant sur des régions de séquence les plus prometteuses.
Banques à haute diversité, construites
de manière experte pour votre cible
Les méthodes de construction de banques sur mesure traditionnelles peuvent produire des codons redondants, une diversité imprévisible ainsi qu’une distribution d’acides aminés non uniforme.
Avec une précision base par base, notre plateforme de synthèse d’ADN sur silicone permet un contrôle précis de la diversité des banques, du rapport de distribution des acides aminés, de la variation de longueur des CDR, ainsi que de l’utilisation des codons, ce qui permet la construction de banques sur mesure de haute qualité, se concentrant sur des régions de séquence les plus prometteuses.

Un moteur de recherche personnalisable, adapté à vos besoins
Concentrez-vous sur la banque et les régions de séquence de votre choix
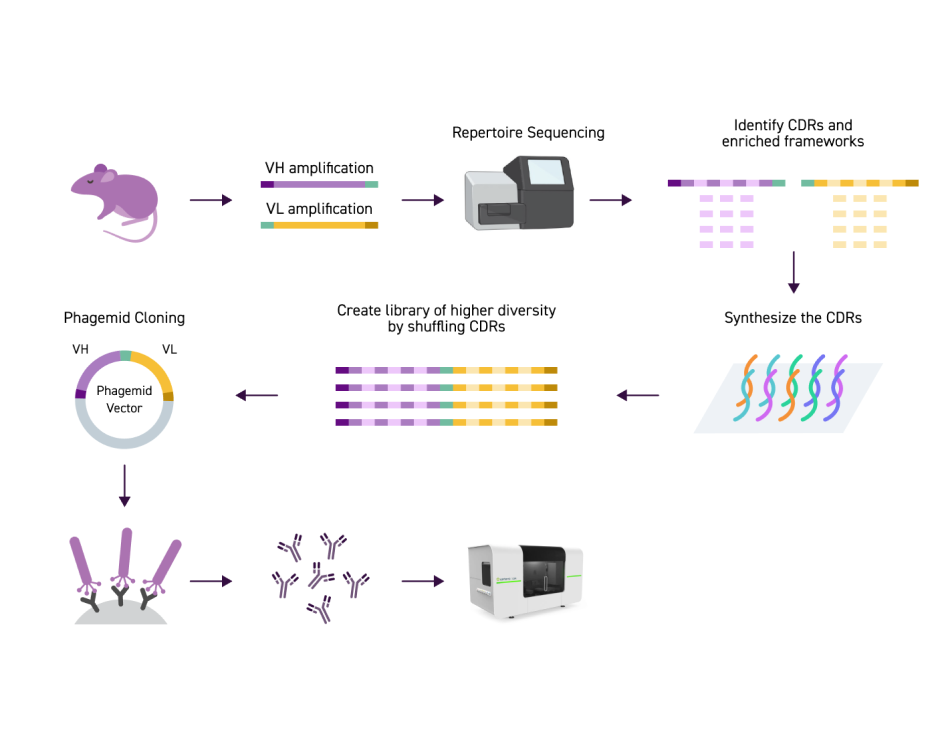
Nous permettons à nos partenaires d’atteindre leurs objectifs en matière de R&D avec une efficacité inégalée grâce à une approche de banque sur mesure à multiples facettes. À partir de différents points de départ (par ex. immunisations, PBMC, bases de données, etc.), nous pouvons construire des variants combinatoires immunisés ou élaborer des banques (VH/VL) adaptées à votre programme de recherche d’anticorps. Nos banques de substitution binaires offrent une humanisation précise en minimisant les risques d’immunogénicité tout en optimisant le potentiel thérapeutique.
Nous proposons également des banques de substitution employant une méthode traditionnelle influençant le rapport en faveur des acides aminés de type sauvage, ce qui limite la complexité à chaque position de variant de même que les substitutions combinatoires à partir du type sauvage. Pour ceux qui nécessitent une encore plus grande précision, notre banque de substitution individuelle permet des compositions définies par l’utilisateur avec des substitutions d’ordre plus élevé basées sur la diversité du domaine.

Concentrez-vous sur la banque et les régions de séquence de votre choix
Nous permettons à nos partenaires d’atteindre leurs objectifs en matière de R&D avec une efficacité inégalée grâce à une approche de banque sur mesure à multiples facettes. À partir de différents points de départ (par ex. immunisations, PBMC, bases de données, etc.), nous pouvons construire des variants combinatoires immunisés ou élaborer des banques (VH/VL) adaptées à votre programme de recherche d’anticorps. Nos banques de substitution binaires offrent une humanisation précise en minimisant les risques d’immunogénicité tout en optimisant le potentiel thérapeutique.
Nous proposons également des banques de substitution employant une méthode traditionnelle influençant le rapport en faveur des acides aminés de type sauvage, ce qui limite la complexité à chaque position de variant de même que les substitutions combinatoires à partir du type sauvage. Pour ceux qui nécessitent une encore plus grande précision, notre banque de substitution individuelle permet des compositions définies par l’utilisateur avec des substitutions d’ordre plus élevé basées sur la diversité du domaine.
Un moteur de recherche personnalisable, adapté à vos besoins
Accès à des banques sans contrainte et sans risque
Afin de vous aider à éviter les problèmes en aval, nous supprimons toutes les séquences d’acides aminés associées aux capacités de développement et aux complications liées à la fabrication. Pour toutes nos banques sur mesure, nous pouvons supprimer de manière sélective des motifs associés à la glycosylation, à l’asparagine désamidation, à l’aspartate isomérisation, à la lysine glycation, à la liaison CD11c/CD18, à la fragmentation, ainsi qu’à l’hydrophobicité.
Des pools d'oligonucléotides peuvent être conçus de façon à correspondre au répertoire CDR naturel
Les contraintes peuvent être supprimées, par ex. isomérisation, clivage, désamidation, sites de glycosylation
Échantillonnage rationnel à partir des régions de séquence souhaitées
Représentation précise, par ex. les séquences de motifs peuvent être explicitement encodées en oligonucléotides
Validation par NGS pour un meilleur CQ
Chacune de nos banques sur mesure est validée par un séquençage de nouvelle génération (NGS) afin de déterminer les variants présents avant la sélection. Nous veillons à obtenir une couverture supérieure à 105 valeurs pour assurer une incorporation de toute la diversité ainsi qu’une distribution uniforme des variants.
Accès à des banques sans contrainte et sans risque
Afin de vous aider à éviter les problèmes en aval, nous supprimons toutes les séquences d’acides aminés associées aux capacités de développement et aux complications liées à la fabrication. Pour toutes nos banques sur mesure, nous pouvons supprimer de manière sélective des motifs associés à la glycosylation, à l’asparagine désamidation, à l’aspartate isomérisation, à la lysine glycation, à la liaison CD11c/CD18, à la fragmentation, ainsi qu’à l’hydrophobicité.
Des pools d'oligonucléotides peuvent être conçus de façon à correspondre au répertoire CDR naturel
Les contraintes peuvent être supprimées, par ex. isomérisation, clivage, désamidation, sites de glycosylation
Échantillonnage rationnel à partir des régions de séquence souhaitées
Représentation précise, par ex. les séquences de motifs peuvent être explicitement encodées en oligonucléotides
Validation par NGS pour un meilleur CQ
Chacune de nos banques sur mesure est validée par un séquençage de nouvelle génération (NGS) afin de déterminer les variants présents avant la sélection. Nous veillons à obtenir une couverture supérieure à 105 valeurs pour assurer une incorporation de toute la diversité ainsi qu’une distribution uniforme des variants.
Contactez-nous
Nos experts en anticorps se tiennent à votre disposition et aimeraient en savoir plus sur vos besoins en matière de recherche. Les cibles difficiles sont les bienvenues !