Using Massively Parallel Reporter Assays (MPRAs) to elucidate the function of genetic variation
Molecular biology has seen a windfall of data over the past 15 years thanks to the proliferation of Next Generation Sequencing (NGS) technology. Genetic data of unprecedented detail can now be gathered on a population scale and leveraged for pharmacogenomics, personalized medicine, preventative healthcare, and to advance our understanding of basic genetics.
To realize the potential of this data researchers must first be able to elucidate the impact of genetic changes. When changes are found in protein-coding regions, the effect can be predicted based on known codon sequences. If the changes occur in non-protein-coding regions, however, the analysis becomes much more difficult.
98% of the human genome consists of non-protein-coding DNA, much of which has unknown functions. Nonetheless, noncoding DNA houses important genetic elements that can alter gene expression levels through genetic and epigenetic means, such as enhancers, silencers, promoters, and other so-called regulatory elements. The average protein-coding gene is governed by multiple non-coding regulatory elements whose activity can vary in a context-dependent manner, making it exceedingly difficult to both identify these elements and link them to the transcriptional activity of specific genes.
Without a detailed understanding of these regulatory elements, potentially clinically relevant information coded within the growing influx of genetic data will remain undiscovered.
To help convert data into knowledge, researchers are turning to Massively Parallel Reporter Assays (MPRAs).
What is a Massively Parallel Reporter Assay?
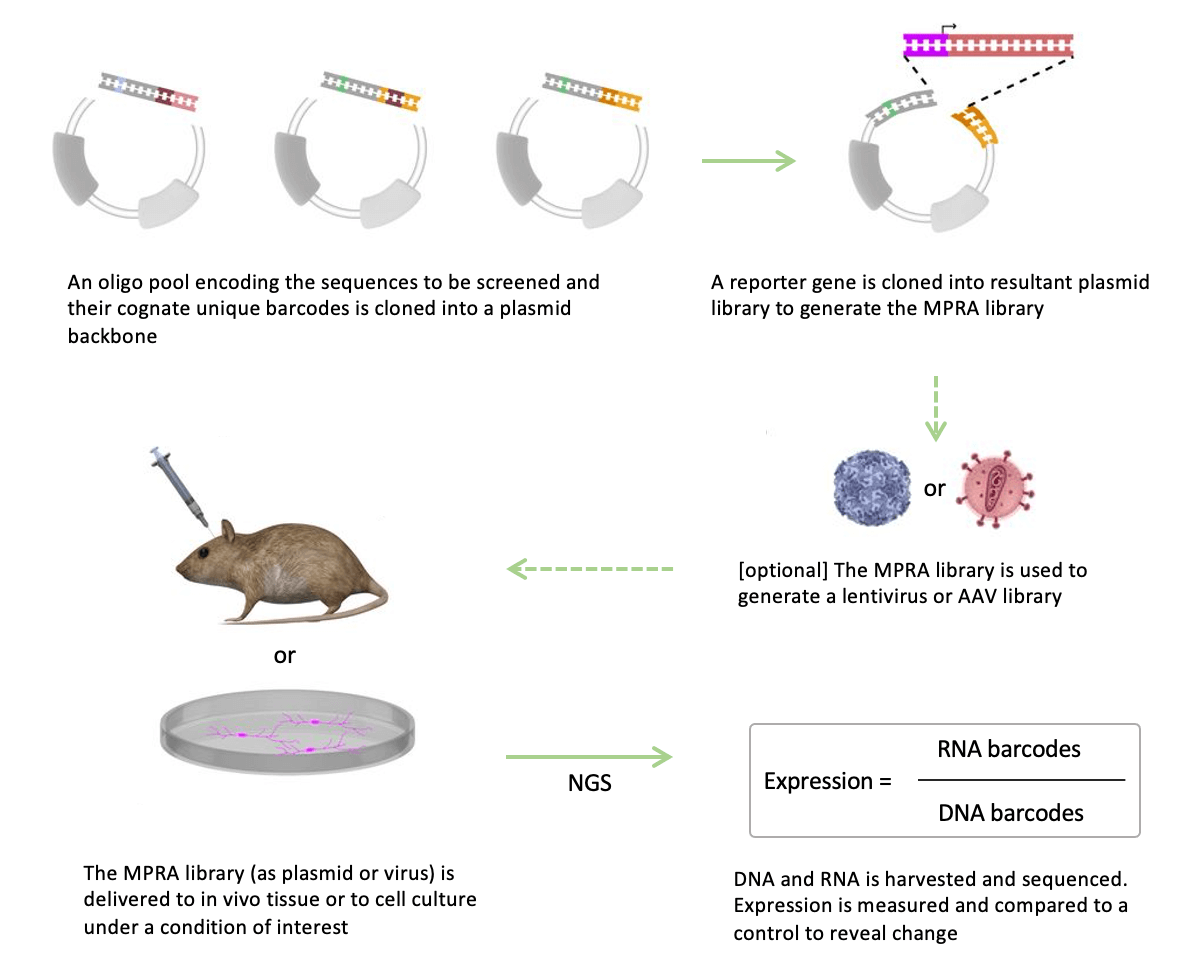
The MPRA is a technique used in molecular biology to simultaneously interrogate the activity of multiple candidate genetic regulatory elements.
At its core, MPRAs are an evolved reporter assay. Simple reporter assays involve cloning a specific regulatory element of interest, such as an enhancer or promoter, into a plasmid upstream of a reporter gene, often luciferase. The plasmid construct is then transfected into a cell line and the resulting luminescence is measured to learn about the transcriptional influence of the regulatory element under varying conditions. This can also be used to determine if a genetic variant alters the activity of a regulatory element.
MPRAs apply the traditional reporter assay at a massive scale, allowing researchers to study several thousand different genetic regulatory elements at once. As MPRA expert Dr. Joseph Dougherty of Washington University in St. Louis explains, “Once you get up to challenges of human genetics, where you identified 100 different loci that are associated with disease, each of which has several hundred associated genetic and epigenetic variants, you’re talking about thousands and thousands of constructs in cells.”
At this scale, the simple luciferase-based reporter assay is not adequate.
MPRAs, though, enable the investigation of many different regulatory elements within a single cell. This is achieved by adding genetic barcodes to the reporter gene. Instead of measuring fluorescence as a readout, researchers can perform RNA-seq to determine which regulatory elements were active and how many transcripts of their respective reporter genes were made as a result (with each one identifiable by its unique barcode).
What to consider when synthesizing a reporter library for MPRAs
Designing a successful MPRA requires the precise synthesis of many thousands of regulatory elements and their associated barcodes. But precision is not easy to achieve with traditional oligonucleotide synthesis. And, interrogating regulatory elements often requires long oligonucleotides that incorporate multiple possible regulatory sequences. With the poor fidelity of array-based oligo synthesis leading to high error rates and biased reporter libraries, MPRAs relying on such libraries can produce misleading results.
To avoid this and maximize the utility of MPRAs, high-quality and precise oligonucleotide synthesis is required.
Twist Bioscience specializes in producing exactly that—accurate, high-quality oligonucleotides for myriad applications, including MPRA.
Accurate, uniform, and long oligo libraries for MPRA
When synthesizing a large-scale reporter library, it’s important to pay close attention to the accuracy, uniformity, and length of oligonucleotides being made.
Twist Bioscience has developed a silicon-based DNA synthesis platform that allows rapid, large-scale production of accurate and highly uniform libraries of oligos up to 300 base pairs in length. These libraries have many advantages for MPRAs:
- Accurate oligonucleotide synthesis allows you to have confidence in your results and trust that the DNA sequence you design is the sequence you’ll test;
- Starting with a highly uniform library removes bias from over and underrepresented elements; and,
- Having access to longer oligonucleotide libraries affords researchers a chance to include barcodes without sacrificing the sequence of interest, as well as more of the natural genetic context in regulatory elements—potentially enabling more accurate results.
For these reasons, Dr. Dougherty’s lab and many others use Twist Bioscience when performing MPRAs. To learn more about how Dr. Dougherty is using Twist Oligo Pools, check out his lab’s recent preprint article here. Similarly, researchers at the Research Institute of Molecular Pathology in Vienna, Austria recently spoke in a webinar about decoding transcriptional regulation using MPRAs and Twist Oligo Pools.
If you're interested in learning more about Twist's oligonucleotide pools, their applications, or simply want to find more resources, visit our Oligo Pools page.
¿Qué piensa?
Me gusta
No me gusta
Me encanta
Me asombra
Me interesa