Designer Enzymes with Beneficial Properties Generated Using Twist Bioscience Genes

Designing new enzymes is an important pursuit for biotechnology. A recent publication in the journal Nature has succeeded in building new, functional protein sequences from the ground-up. In the publication, the researchers developed an automated tool that built new proteins out of a library of new modular protein parts. Then, using Twist Bioscience gene synthesis, they generated and tested these new proteins in the lab. Here, we examine this groundbreaking study and see how it has impacted the future of enzyme engineering.
Proteins are essential to life. Enzymes (one kind of protein) act as catalysts, conducting chemical reactions that are central to metabolism, detoxification, and the control of cellular function. Enzymes are able to react on a broad range of substrates, and individual enzymes are often highly specific for certain chemical structures. Additionally, some enzymes conduct reactions that produce useful and unusual chemical structures that would be difficult to access using traditional chemistry. These properties have made enzymes a cornerstone of both synthetic biology and biotechnology.
By using individual, or designed cascades of enzymes, desirable chemical reactions can be conducted either within cells or simply in a test tube. Scientists have been able to use enzyme-driven techniques to convert low-value input molecules like farm-waste cellulose into high-value products like flavours, scents, pharmaceuticals, biofuels and oil-replacement plastics.
An enzyme’s activities, namely its substrate specificity and its reaction rate, are dictated by its amino acid sequence. The amino acid sequence also controls the enzyme’s 3D folded structure. However, while the natural repertoire of enzymes is vast (conducting over 10.000 known reactions) natural proteins are, naturally, optimal for their function in nature and not in a laboratory. This can lead to issues with poor enzyme stability, slower-than-expected reaction speeds and weak activity on non-natural chemicals. Such properties often lead to natural enzymes being unsuitable for industrial scale biotechnology. Scientists have therefore conducted significant research on enzyme engineering, in which the amino acid sequence of a protein is modified to optimize its properties.
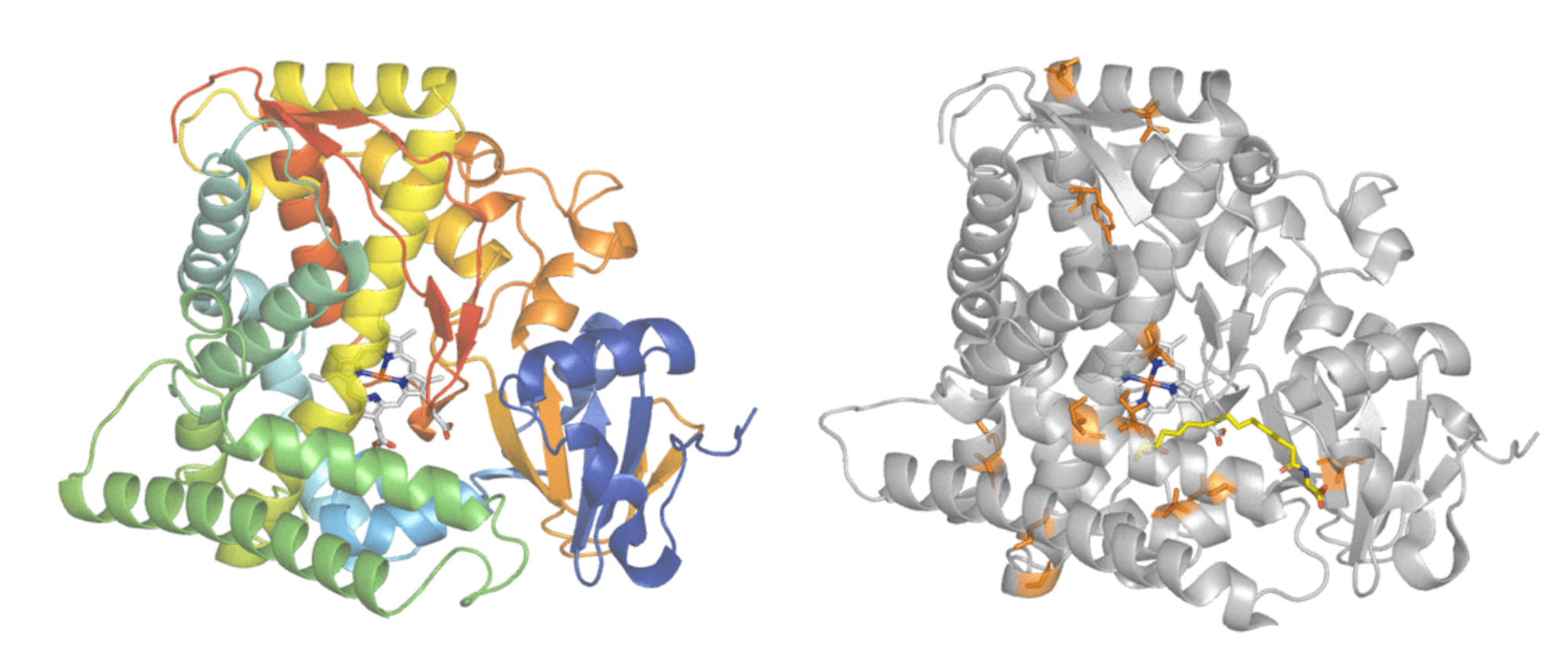
Protein engineering is commonly used to improve enzyme features. Cytochrome P450BM3 (left) is responsible for adding oxygen atoms to long fatty acid molecules. An engineered form of Cytochrome P450BM3 (right) had 11 amino acid changes made (highlighted in orange) to give it activity on the industrially useful, non-natural compound iodomethane. Protein structures - PDB: 1FAH and PDB: 3CBD. Images modified in PyMol.
One highly desirable, but so far elusive application in enzyme engineering is the design and construction of entirely new enzyme sequences from the ground-up. There are no universal rules defining the sequence-structure-function relationship of enzymes, making it almost impossible to design a sequence that can fold into a molecule of desired function. However, the ability to make new enzymes in such a way would allow researchers to do away with everything superfluous in natural enzymes, sticking to just structures required for both catalysis and stability in a given condition. In theory, this will allow for highly efficient bioconversions on any desired molecule, making enzymes far more tractable to the chemical industry.
A recent paper published in Nature by Gideon Lapidoth and colleagues of the Weizmann Institute of Science in Israel, entitled “Highly active enzymes by automated combinatorial backbone assembly and sequence design,” takes steps towards this ultimate enzyme design goal. The study describes the design and implementation of an automated method that allows researchers to generate new, non-natural, but functional enzyme sequences from sequences describing known enzyme folds.
To give some more background to the study, the research focuses on a protein architecture called a TIM-barrel. TIM-barrel proteins look similar to microscopic Life Savers candies when folded into their characteristic hollow circular shape. This protein structure is remarkable, as the TIM-barrel fold is thought to be one of the earliest complex protein structures to have evolved in nature. As a result it is present in almost 10% of all proteins.
TIM-barrel folds are found in five of the six main superfamilies of enzymes, consisting of 70 distinct sequence families, each with very diverse amino acid sequences and reactivity. It is also known that in many TIM-barrel proteins, the amino acids that controlling TIM-barrel structure and the amino acids that control activity are distinct. Therefore, in theory, this property makes TIM-barrels an ensemble of modular parts, some structural and some functional. It should also be possible to take catalytic residues from one TIM-barrel, and swap into another TIM-barrel to produce new and interesting enzymes.
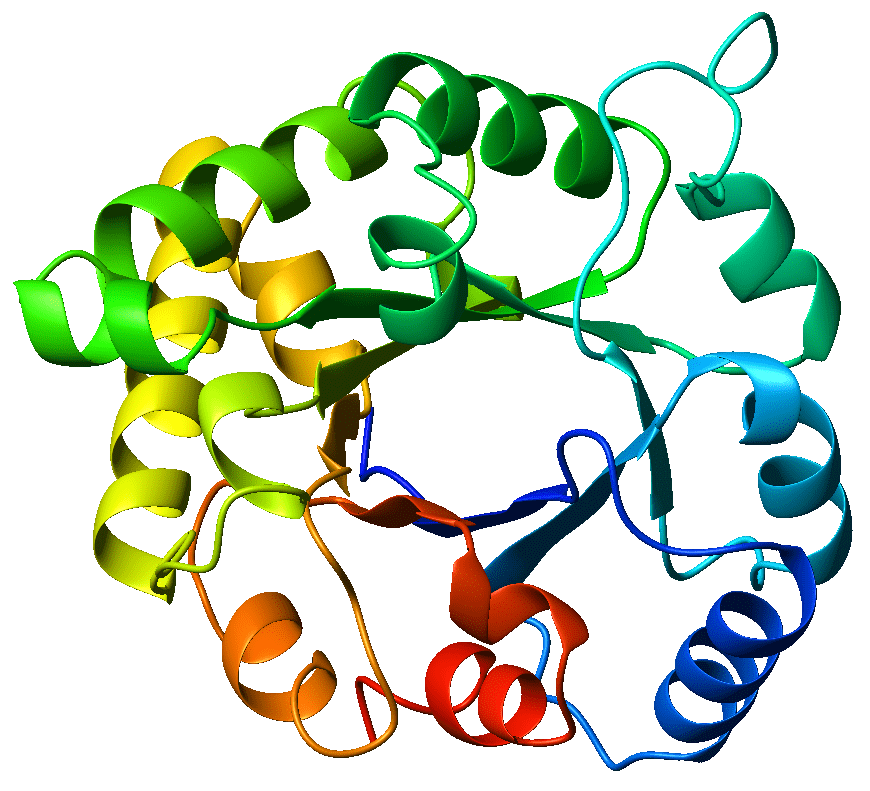
The circular crystal structure of the TIM-barrel enzyme triosephosphate isomerase - this is the enzyme from which TIM-barrels get their namesake. Fuente: Wikicommons. Author: WillowW. Image licensed under CC BY-SA 3.0.
By exploiting this property, this study aimed to design new TIM-barrels based on two distinct families of the protein (the glycoside hydrolase 10 xylanases and the phosphotriesterase lactonases). The researchers developed an automated method that initially makes a library of modular TIM-barrel parts belonging to both families. New TIM-barrel backbone structures containing the same catalytic residues as the natural enzymes can then be constructed from this library of parts. In order to maximize the chance of generating functional proteins, the method maximizes the compatibility between parts, and incorporates known stabilizing mutations. Optimal selection of the modular parts to form new backbone structures was also informed by another algorithm originally designed to engineer antibody sequences, called Rosetta. The final result was 77 new TIM-barrel protein designs from two families, generated from the ground-up by a computer.
When it came to testing their designs, as the designed sequences had never existed in nature before, the team turned to Twist Bioscience’s Gene Synthesis Platform to synthesize the DNA sequences that encoded their novel TIM-barrels. Strikingly, of the 77 designs, each and every one could be introduced and expressed in E. coli, suggesting that despite being designed by a computer, the proteins were able to fold into a protein-typical globular structure.
Of the 77 proteins, 28 were also found to be active, 21 showed xylanase activity, and seven showed lactonase activity. Additionally, four of the functional proteins showed activities that are comparable to their natural counterparts. These results provide the first convincing evidence that the design of functional, useful TIM-barrel enzymes from the ground up is possible when using libraries of modular TIM-barrel parts.
Importantly, this study opens up doors for further research into protein design. It now allows researchers to begin exploring how each modular part contributes to enzyme function, bridging the gap between the structure-function relationship in TIM-barrel proteins. In the article, the researchers suggest that their results will assist with further research that builds on this design tool, allowing for the engineering of novel TIM-barrel proteins with better activities or even entirely new active site structures, thus leading to new designed activities in the protein family. Additionally, it is no stretch that this study provides a workflow for the design of other protein families in the same manner.
Since Twist Biosciences genes can be delivered cloned into a vector of choice, and verified by NGS as sequence perfect, they are an excellent tool for protein engineering projects like the one described here. This ensures that researchers are satisfied that they are experimenting on their exact designs, even if the designs have not existed in nature before. Additionally, the genes are synthesized on our silicon platform, that can scale to the size of any engineering project. Thousands of unique genes can be made every run, if required. Find out more about Twist Bioscience Gene Synthesis here. We are proud to be an integral part of the rapidly growing protein engineering space, and are excited to see more novel proteins being engineered to provide useful functions in the near future.
¿Qué piensa?
Me gusta
No me gusta
Me encanta
Me asombra
Me interesa
