Twist Bioscience’s Oligo Pools Unlock the Secrets of Non-Coding RNA
The more that scientists peer into the mechanisms controlling how the human genome encodes information and how that information is translated into life, the more beautifully complex the entire system seems. One area of active research is in transcriptional regulation–the forces controlling how and when a protein will be synthesized from genetic information. Better understanding of how the molecules of life are organized and regulated allows us to not only understand what it means to be human on a chemical level, but it also allows us to better identify, anticipate and treat disease. Ongoing research in the field of transcriptomics ultimately allows for the identification of new disease hallmarks, a better understanding of the mechanism behind disease, and ultimately the development of new treatments.
A recent study published in Nature by Dr. Yoav Lubelsky and Dr. Igor Ulitsky of the Weizmann Institute of Science in Rehovot, Israel, used pooled oligonucleotides to understand how a recently discovered molecule called a long non-coding-RNA (lncRNA) can function in transcriptional regulation. They found that specific nucleotide sequences in the lncRNA molecule are responsible for its unusual localization to the nucleus of the cell. The researchers believe their work will enable the “identification of additional sequence and structural elements shared across lncRNAs, and expedite classification of these enigmatic genes into functional families.”
RNA is an enigmatic molecule with a number of functions in the human cell. Messenger RNA (mRNA) is transcribed from our genome, specifically from our genes. Each mRNA molecule is then translated into a protein by a team of RNA and RNA-dependent molecules.
Ribosomes, minute molecular machines consisting of a fusion between ribosomal RNA (rRNA) and variety of ribosomal proteins, create the bonds between the growing protein’s amino acid molecules. Each new amino acid in the protein sequence is encoded by the mRNA and is carried to the ribosomes by another RNA molecule called a transfer RNA (tRNA). There are also RNA molecules that have been discovered with roles in the cell’s DNA replication (for example small nuclear RNA) and transcription regulation (for example small interfering RNA). It is estimated that over 100.000 different functional RNA molecules are produced by any one cell, 5x the number of genes in the genome.
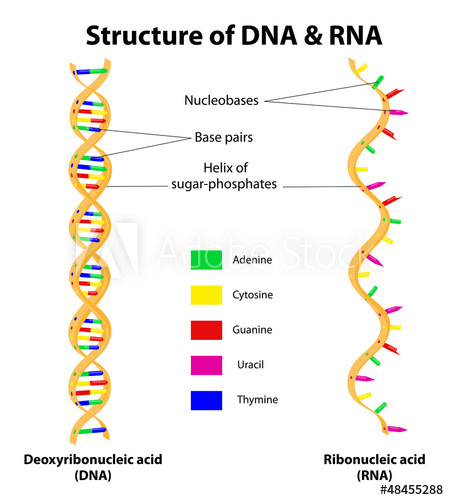
The structure of RNA, a molecule which is transcribed from DNA in the genome.
One little understood RNA molecule in a cell is a family of molecules called long non-coding RNA (lncRNA). LncRNA’s are defined by the following properties:
-
Over 200 nucleic acids long
-
Transcribed from the genome
-
Not protein encoding
-
Found almost entirely in the nucleus of the cell
Thousands of potential lncRNA molecules have been identified. Their main function is the regulation of gene expression with examples both increasing or decreasing the rate of expression dependant on the gene, the lncRNA, and the cellular context.
The fact that most lncRNAs are found in the nucleus is unusual, as many RNA molecules are shuttled out of the nucleus to become proteins or provide other functions. This means there must be something blocking lncRNAs from leaving the nucleus. In their paper, Dr. Lubelsky and Dr. Ulitsky set out to discover the exact nature of this blocking force.
To do so, the researchers conducted a beautiful experiment. They first hypothesized that the sequence of the lncRNA itself has some element facilitating its unusual localization. To test this hypothesis, they took a dataset of 37 lncRNA molecules known to localize to the nucleus, and broke their respective DNA-encoding sequences down into 109 nucleotide long “tiles.” These tiles were then arranged into 5.511 unique sequence combinations, which were synthesised using Twist Bioscience’s high throughput DNA synthesis technology as a pool of high-quality oligonucleotides. These oligonucleotides were then cloned into the front of a gene encoding an mRNA molecule that is known to be readily exported out of the nucleus.
Next, the researchers allowed cells to express this pool of tiled lncRNA-mRNA conjugates. Once expression of the conjugates had taken place, they separated out the nuclei from the cells and used reverse-transcription PCR and next-generation sequencing to investigate if any of the readily exported mRNA molecules had gotten stuck in the nucleus. As per their hypothesis, some of the mRNA molecules were no longer exported and their use of next-generation sequencing allowed them to identify which of the lncRNA tiles were conjugated to these non-exported mRNAs.
Tiles producing the most profound nuclear localization effects contained two common elements which the researcher’s dubbed a “SIRLOIN element”–a sequence that was complementary to a genetic element called an Alu-repeat and a DNA sequence motif of RCCTCCC.
In the genome, an Alu-repeat is another piece in its complex information storage architecture. These are regions of 300 or so base pairs that repeat over and over millions of times to cover up to 10% of the human genome. One of their key functions is in the switching on and off of tissue-specific genes. The researchers suggested that the presence of the complementary Alu-element in lncRNAs has been suggested to allow lncRNAs to base-pair with other Alu-elements in the genome, enriching lncRNAs to the nucleus.
The researchers then used a second pool of oligonucleotides, also synthesized by Twist Bioscience, to investigate the impact of SIRLOIN elements further. The oligo pool consisted of tiles of many other lncRNA SIRLOIN elements, as well as sequences containing modifications to the Alu and RCCTCCC motifs. Again researchers cloned the pool in front of an mRNA that is usually exported from the nucleus.
From studies of this pool, researchers made a number of important discoveries:
-
Mutating the CCTCCC motif to contain purines instead of pyrimidines stops SIRLOIN elements from localizing to the nucleus.
-
They found that an RNA binding protein called Heterogeneous nuclear ribonucleoprotein K (HNRNPK) interacts with SIRLOIN elements.
-
They found that removal of HNRNPK from the cell environment significantly reduced nuclear localization of the SIRLOIN containing mRNAs.
Their observations show that binding of HNRNPK to the SIRLOIN elements is that blocking force keeping lncRNA in the nucleus. It also shows that the binding occurs in a manner dependent on the CCTCCC motif.
This study is exemplary for the use of oligo pools to probe the core mechanisms behind the complexity of life. Being able to access high quality synthetic DNA from Twist Bioscience enabled the high-throughput nature of these experiments. Additionally the low error rates across the oligo pool provided the researchers with the confidence that every sequence in the pool functioned as intended. Furthermore, the uniformity provided in Twist Bioscience’s Oligo Pools allowed the researchers to ensure their experiments weren’t biased toward particular sequences that were tested. While the exact mechanism by which HNRNPK blocks lncRNAs from leaving the nucleus is not yet understood, this study has provided a significant advancement in our knowledge of how some of life’s most enigmatic molecules function in mammalian cells. The researchers believe their work will enable the “identification of additional sequence and structural elements shared across lncRNAs, and expedite classification of these enigmatic genes into functional families.”
Cover image source: Flickr. Author: Joseph Elsbernd. Licensed under CC BY 2.0
¿Qué piensa?
Me gusta
No me gusta
Me encanta
Me asombra
Me interesa
