Advances in DNA Data Storage: Random Access Memory

2018 is already seeing huge advances in the use of DNA as a medium to store data. A new study published by Microsoft Research and researchers at the University of Washington is a breakthrough in storage technology, paving the way for DNA data storage on a scale far beyond traditional storage capabilities. We dive into the details of this study here.
Humans, by their very nature, are archivists. Since the beginning of the digital age, vast troves of digital data have been hoarded. Multinational technology conglomerate Cisco predicted that by 2019, the data that makes up the “Internet of Everything,” (often referred to as the Internet of Things), will surpass a seemingly unfathomable 500 zettabytes (500 sextillion bytes). A data volume that is rapidly inflating due to the wide adoption of personal cloud storage.
However, our world’s data storage capacity is not infinite as it relies on the element silicon which is rarely found pure in nature. By tracking the progression of improvements to data storage density alongside the increase in data stored, researchers predict that by 2040 we will run out of memory-grade silicon on earth.
In anticipation of the “data boom crisis,” researchers and tech companies have been looking for an alternative to long-term data storage. Long-term data is described as archive data that doesn’t need to be accessed often. Currently, long-term data is most often stored in tape archives, and more recently on silicon archives. Tape is non-ideal as it degrades, and needs to be re-copied about once every 10 years. As mentioned, silicon is finite and will eventually be completely mined, which requires researchers to discover a new, more resourceful solution.

A collaboration between the University of Washington and both Microsoft research and Twist Bioscience, announced in Spring 2016, has been focusing on the use of DNA as a data storage device. DNA is nature’s way of storing information about the structure and function of life. With the access to high-throughput, high-quality custom DNA synthesis from Twist Bioscience and the commonality of high throughput next-generation sequencing, life’s data storage medium can in principle be adapted to store digital data too.
Chemically, DNA is an ideal long-term storage solution as it is an incredibly robust molecule, maintaining its integrity for tens of thousands of years when dried out. With current data storage architectures, the highest achieved storage density for synthetic DNA is a massive 215 petabytes per gram. Since the collaboration was announced, new advances in the data storage field have already been funded and published.
In 2016, a vast range of data was stored in small fragments of DNA, including the Universal Declaration of Human Rights translated in 100 languages and a music video by the band OK Go. In 2017, the collaboration announced the first report of archival quality data stored in DNA, preserving two recordings from the Montreux jazz festival which are part of UNESCO’s Memory of the World Archive.
February 2018 marks another huge step in this burgeoning industry. Microsoft and Washington researchers published the first report of DNA-based random access memory (RAM) in the journal Nature. In this study, researchers not only archived record breaking volumes of data in DNA, but they also stored the data in a format that models your device’s RAM for the first time.
Before now, if access to a single DNA-stored file was required, the entire pool of DNA would have to be read by next-generation sequencing. This property is analogous to how a record player can only access the data stored on a vinyl record in sequence—a property that is manageable with small-scale data storage requirements, but doesn’t scale to terabyte volumes.
The device you are using to read this article contains RAM hardware, which can access any part of the stored data instantaneously, regardless of how or where the data is stored. As you start a program on your device, all the files that the program runs from are loaded onto the device’s RAM alongside all of the data the program is manipulating. As RAM data is accessed instantaneously, any part of that program can be run or computed at a clicks notice. As a result, your experience using your device is typically seamless.
The authors of the study hypothesized that if archival DNA could be synthesized in a way that allows a decoder to treat it as RAM and not as a sequential storage device, single files could be accessed quickly without ever having to sequence the entire DNA pool.
The Polymerase Chain Reaction (PCR) is an enzymatic method that allows for the exponential amplification of a target DNA sequence. To target a sequence, two small single-stranded DNA sequences called primers are designed. The primers flank the sequence for amplification, and the enzymatic process photocopies the flanked region.
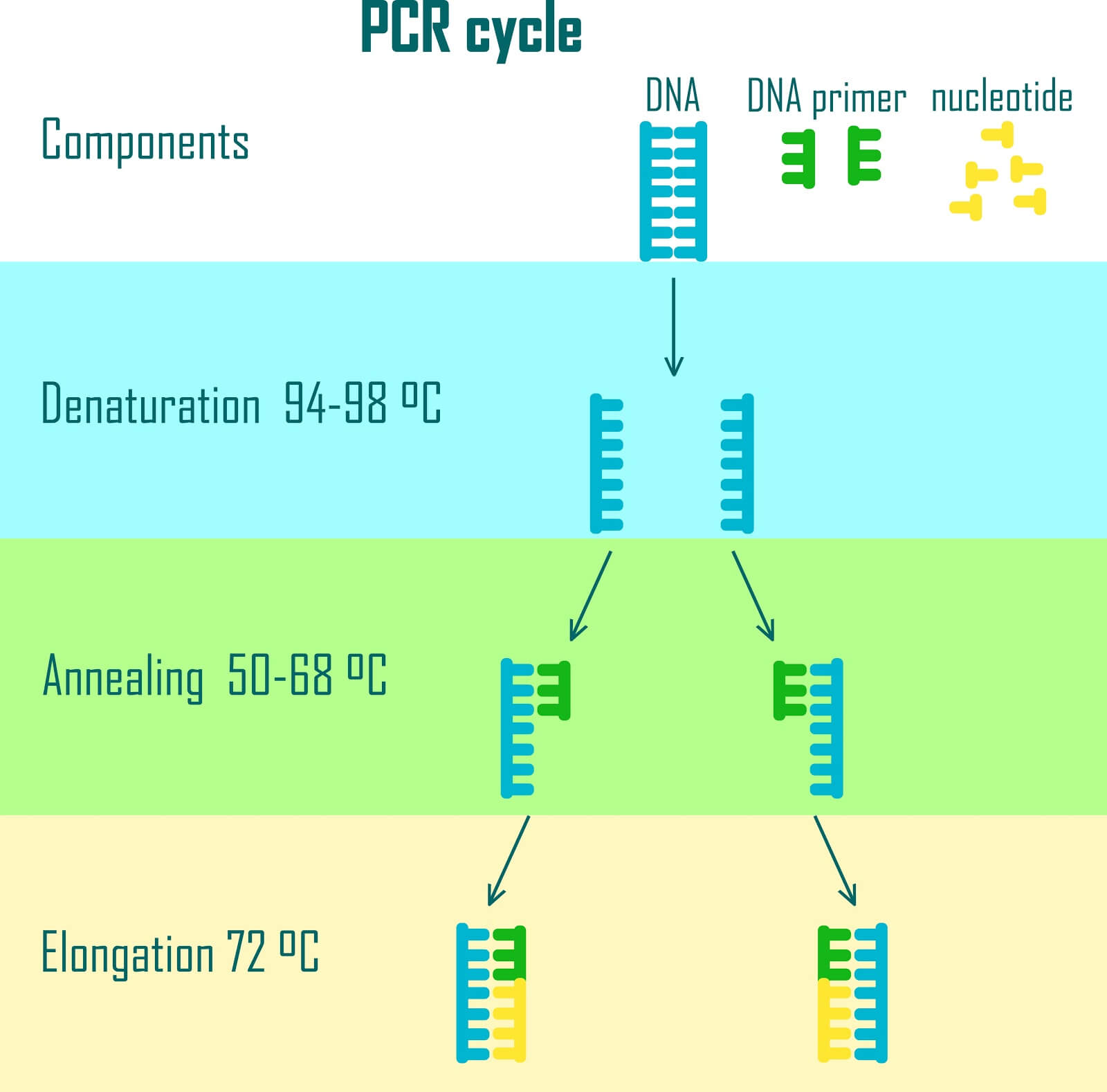
During the step where sequences are designed to encode data, additional sequences are also added to either side of each encoded file. Primers are then designed to match these flanking sequences and amplify out a single file from a pool of DNA.
By flanking each file with a unique sequence corresponding to different primers, any file can be accessed independently from the pool. In a device, the data stored in RAM is treated in an analogous way in a process called memory addressing .
Using this memory architecture in DNA, the research team then encoded 200 MB of data in 35 files. The data was then stored in almost 13.5 million unique, high-quality DNA sequences all around 150 bp in length, each synthesized by Twist Bioscience. Twist Bioscience leads the market in high throughput, low-cost DNA synthesis. The inherent scalability of a silicon platform provides a path to the scales required for viable DNA data storage. The files stored included, amongst other things, an HD version of the band OK Go’s music video for their song This Too Shall Pass and a collection of classical music.
The authors were then able to select individual files or groups of files, use PCR to amplify these files from the pool of DNA, and then use a MinION DNA sequencer to sequence the amplified files. The sequencing data was then decoded to recover every file byte-for-byte without error!
In the study, the authors comment that this is one of the first studies that encodes data in a way that is truly scalable to the petabyte scale, ready for massive long-term data storage in DNA. With current trends in improvements to DNA synthesis efficiency, leading to decreases in the cost per base, DNA data storage is set to move beyond just proof-of-principal in the near future. Humanity is already preparing for the upcoming data-crisis by finding innovative materials to store data. With such technologies, the predicted upcoming data-crisis may never come to pass.
What did you think?
Like
Dislike
Love
Surprised
Interesting